THE XMM-NEWTON ABC GUIDE, STREAMLINED
RGS, Hera Parameter Window
Contents
Prepare the Data
Reprocess the Data
Make Images
Make a Light Curve
Generate and Apply a New GTI
Create the Photon Redistribution Matrix (RMF)
Check for Pile Up
Combining Spectra
Prepare the Data
Please note that the two tasks in this section (cifbuild and odfingest) must be run in the ODF directory. These are the only tasks with that requirement, and after this section, we will work exclusively in our reprocessing directory.Many SAS tasks require calibration information from the Calibration Access Layer (CAL). Relevant files are accessed from the set of Current Calibration File (CCF) data using a CCF Index File (CIF). To make the ccf.cif file, navigate into the ODF directory using the icons in the User Account Window of the Hera interface. Then, in the Tool Parameter Window, in the Task Name box, enter cifbuild and click "Get".
The task odfingest extends the Observation Data File (ODF) summary file with data extracted from the instrument housekeeping data files and the calibration database. It is only necessary to run it once on any dataset, and will cause problems if it is run a second time. If for some reason odfingest must be rerun, you must first delete the earlier file it produced. This file largely follows the standard XMM naming convention, but has SUM.SAS appended to it. To run odfingest, in the Tool Parameter Window, in the Task Name box, enter odfingest and click "Change".
Hera automatically resets the relevant environmental parameters to the output of these tasks, so we can continue merrily on our way.
Reprocess the Data
To reprocess the data, go up one directory in the tree and make a new working directory using the buttons in the upper left corner of the User Account Window. When you are in the working directory, call rgsproc. As in the previous section, just enter the task name in Task Name box and click "Change". The default parameters are fine for most datasets, so click "Run rgsproc".This takes several minutes, and outputs 12 files per RGS, plus 3 general use FITS files. It is convenient to rename them something easy to type; this can be done by clicking on the pen icon next to the file name in the User Account Window. We'll assume the new name for the RGS1 and RGS2 event files are r1_evt1.fits and r2_evt1.fits, respectively. The pipeline task rgsproc is very flexible and can address potential pitfalls for RGS users. If the default parameters are sufficient for your data (and they should be for most), feel free to skip ahead. However, there are some cases where the standard processing may not be appropriate. These are discussed next.
First, if the data includes a nearby bright optical source, with certain pointing angles, zeroth-order optical light may be reflected off the telescope optics and onto the RGS CCD detectors. If this falls on an extraction region, the current energy calibration will require a wavelength-dependent zero-offset. Stray light can be detected on RGS DIAGNOSTIC images taken before, during and after the observation. This test, and the offset correction, are not performed on the data before delivery. To check for stray light and apply the appropriate offsets, enter rgsproc in the Task Name box and click "Change". Then,
- Set the calcoffsets parameter to yes.
- Click "Run rgsproc".
If the data includes a nearby bright X-ray background source that is well-separated from the target in the cross-dispersion direction, a mask can be created that excludes it from the background region. Here the source has been identified in the EPIC images and its coordinates have been taken from the EPIC source list which is included among the pipeline products. The bright neighboring object is found to be the third source listed in the sources file. The first source is the target, enter rgsproc in the Task Name box and click "Change". Then,
- Set the withepicset parameter to yes. Set the epicset parameter to name of the EPIC source list, such as what is generated by emldetect or eboxdetect; it will have the general form P*EMSRLI*FTZ. Set exclsrcsexpr to INDEX==1&&INDEX==3.
- Click "Run rgsproc".
If the true coordinates of an object are not included in the EPIC source list or the science proposal, the user can define the coordinates of a new source. Enter rgsproc in the Task Name box and click "Change". Then,
- Set withsrc to yes and srclabel to the source name, Mkn421. Set srcra and srcdec to the RA and Declination, respectively, both in decimal degrees: 166.113808 and +38.208833.
- Click "Run rgsproc".
Make Images
Two commonly-made plots are those showing PI vs. BETA_CORR (also known as "banana plots") and XDSP_CORR vs. BETA_CORR.To make a banana plot, enter evselect into the Task Name box and click "Change". Then,
- By the table parameter, use the Browse button to select the event file, r1_evt1.fits.
- Set withimageset to yes. This will make parameters that are specific to making an image appear.
- Enter an output file name in imageset. We will use pi_bc.fits. Set xcolumn to BETA_CORR and ycolumn to PI. In the expression parameter box, enter (FLAG == 0)&&(PI in [300:2000]).
- Set imagebinning to imageSize, ximagesize to 600, yimagesize to 600.
- Click "Run evselect".
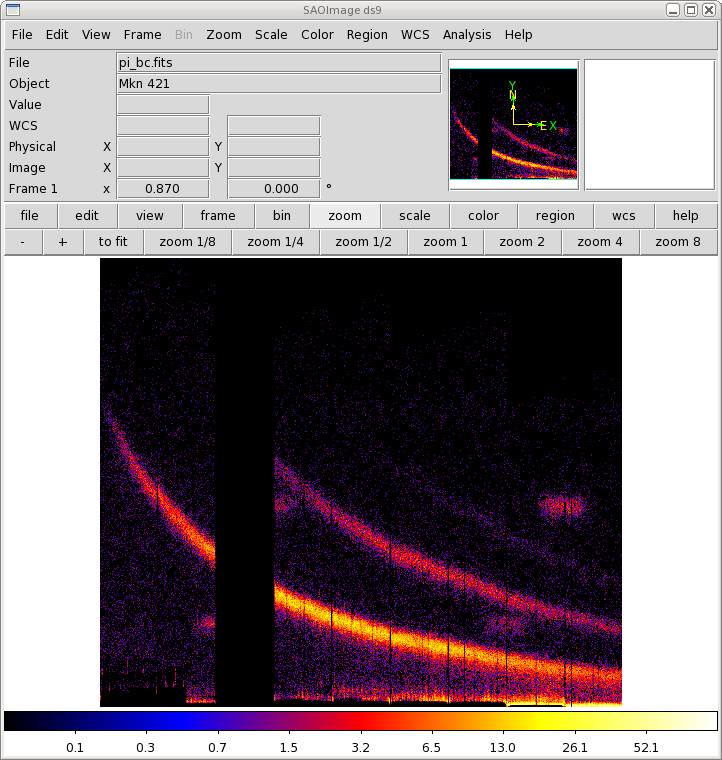
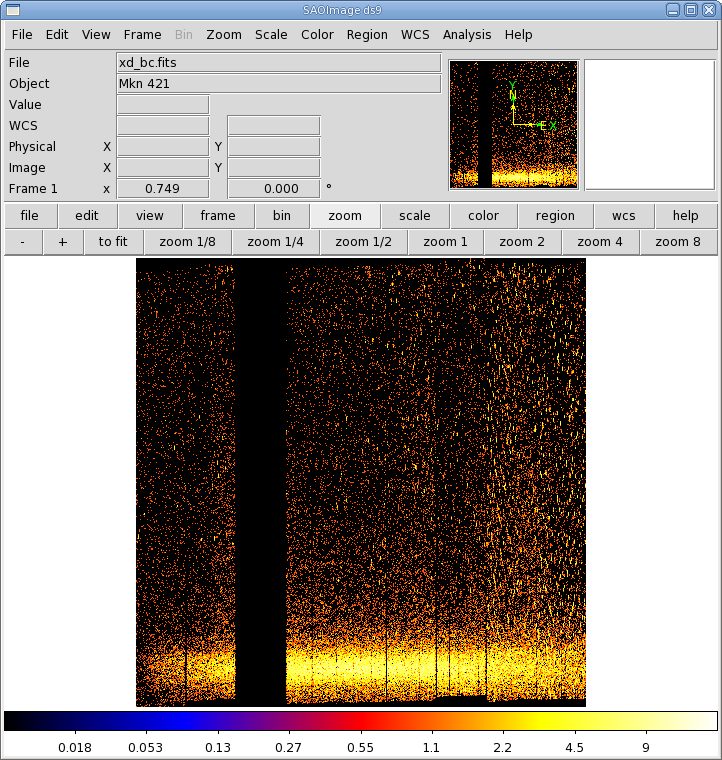
|
Make a Light Curve
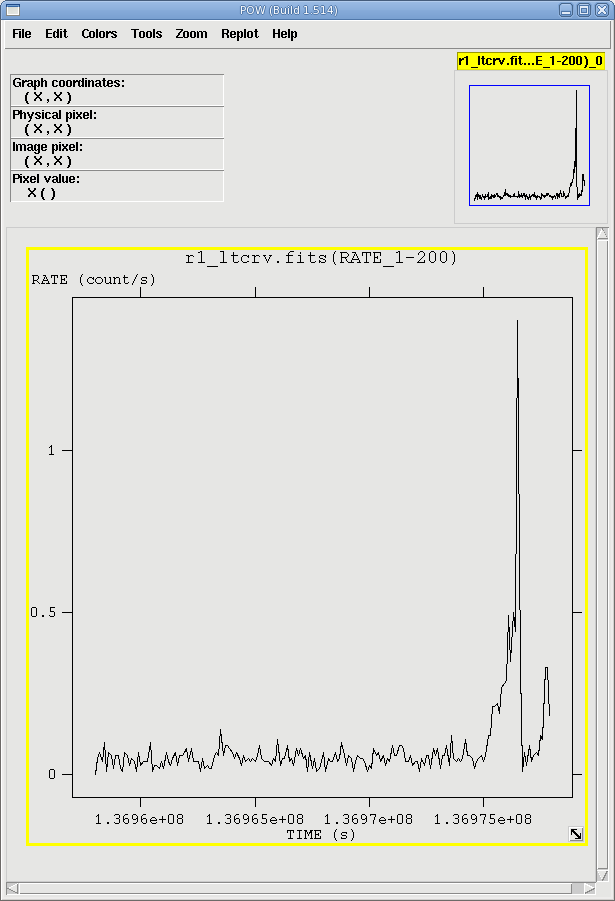
The XMM-Newton Observatory is susceptible to soft particle flaring, so it is necessary to examine the light curve to determine how much of the data is useful. We will extract a region, CCD9, that is most susceptible to these events and generally records the least source events due to its location close to the optical axis. Also, to avoid confusing background for source variability, a region filter that removes the source from the final event list should be used. The region filters are kept in the source file product P*SRCLI*.FIT.More experienced users should be aware that with SAS 13, the *SRCLI* file's column information changed. rgsproc now outputs an M_LAMBDA column instead of BETA_CORR, and M_LAMBDA should be used to generate the light curve. (The *SRCLI* file that came with the PPS products still contains a BETA_CORR column if you prefer to use that instead.)
To create a light curve, enter evselect into the Task Name box and click "Change". Then,
- By the table parameter, use the Browse button to select the event file, r1_evt1.fits.
- Set withrateset to yes. This will make the parameters that are relevant specifically to making a light curve appear.
- Set maketimecolumn to yes, set timebinsize to 100, and enter an output file name in rateset. We will use r1_ltcrv.fits. In the expression parameter box, enter (CCDNR==9)&&(REGION(P0153950701R1S001SRCLI_0000.FIT:RGS1_BACKGROUND,M_LAMBDA,XDSP_CORR)).
- Click "Run evselect".
The output file r1_ltcrv.fits can be download and viewed with fv, as shown
in Figure 2.
- fv r1_ltcrv.fits &
Generate and Apply a New GTI
Examination of the lightcurve shows that there is a noisy section at the end of the observation, after 1.36975e8 seconds, where the count rate is well above the normal background count rate of about 0.05 count/second. There are two procedures that make the GTI file (gtibuild and tabgtigen) that, when applied to the event file in another run of rgsproc, will excise these sections.The first method, using gtibuild, requires a text file uploaded from your local machine as input. In the first two columns, refer to the start and end times (in seconds) that you are interested in, and in the third column, indicate with either a + or - sign whether that region should be kept or removed. Each good (or bad) time interval should get its own line. In the example case, we would write in our ASCII file gti.txt:
1.36958e8 1.36975e8 +Upload the file by browsing your local directory in the User Account Window, and, in the Task Name box, enter gtibuild. Then,
- For the file parameter, enter gti.txt. In the table parameter, enter the output file name; we will use gti.fits.
- Click "Run gtibuild".
We can apply the GTI to the event file by running rgsproc again. rgsproc is a complex task, running several steps, with five different entry and exit points. It is not necessary to rerun all the steps in the procudure, only the ones involving filtering. Enter rgsproc in the Task Name box and click "Change". Then,
- Set entrystage to 3:filter and finalstage to 5:fluxing. Set auxgtitables to the new gti file, gti.fits.
- Click "Run rgsproc".
Create the Photon Redistribution Matrix (RMF)
Response matrices (RMFs) are not provided as part of the pipeline product package, so you must create your own before analyzing data. The task rgsproc generates a response matrix automatically (with the nomenclature *RSPMAT*.FIT), but as noted above the source coordinates are under the observer's control. The source coordinates have a profound influence on the accuracy of the wavelength scale as recorded in the RMF that is produced automatically by rgsproc, and each RGS instrument and each order will have its own RMF.
Making the RMF is easily done with rgsrmfgen. Please note that, unlike with EPIC data, it is not necessary to make ancillary response files (ARFs).
To make the RMFs, enter rgsrmfgen in the Task Name box and click "Change". Then,
- By the table parameter, use the Browse button to select the event file r1_evt2.fits. In a similar way, select the spectrum file, P0153950701R1S001SRSPEC1001.FIT. In the rmfset parameter, enter the output file name; we will use r1_o1_rmf.fits.
- Set the number of energy bins, rows, to a number greater than 3000; we will use 5000. Set the lower and upper energy limits of the response file by setting emin to 0.4 and and emax to 2.5.
- Click "Run rgsrmfgen".
At this point, the spectra can be either analyzed or combined with other spectra. However, if your source is very bright (total count rate in all orders > 12 cts/s in RGS1, and 6 cts/s in RGS2), it is a good idea to check it for pile up first.
Check for Pile Up
Depending on how bright the source is event pile up may be a problem. Pile up occurs when a source is so bright that incoming X-rays strike two neighboring pixels or the same pixel in the CCD more than once in a read-out cycle. In the RGS, it will cause two coincident 1st order photons to combine into a single 2nd order event at the same spatial position on the detector but at half the wavelength; this results in photons moving from 1st order to 2nd order, and 2nd order to 3rd. Further, it can increase the number of complicated event patterns of the sort that the on-board processing removes; this results in differences in pile up between the two RGS detectors, and between CCD locations.One way to determine if there is pile up is to examine the ratio of 1st and 2nd order fluxed spectra. Non-piled sources should have fluxed spectra that agree within a few percent, while piled sources will show discrepancies of about 10% or higher.
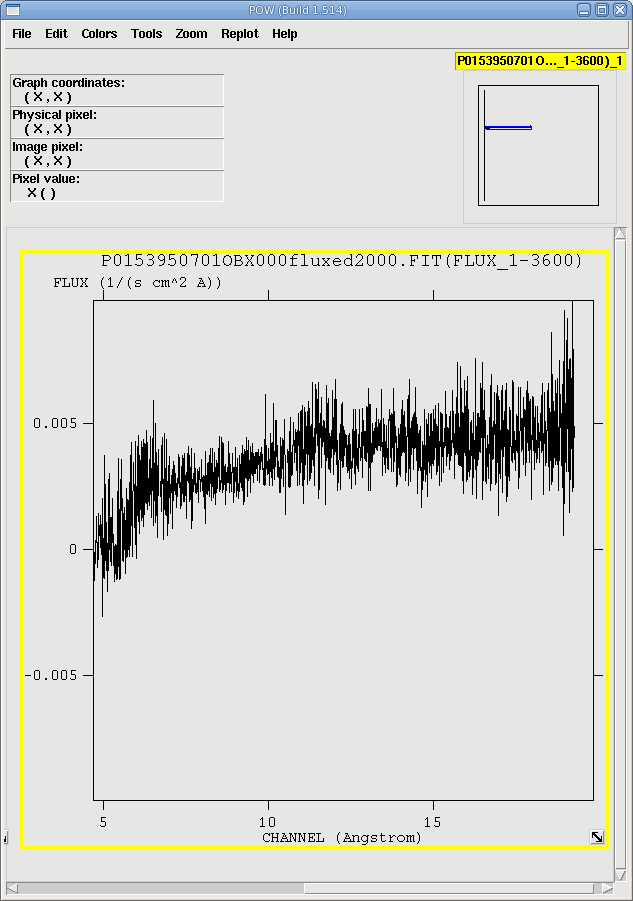
When rgsproc is run through the 5:fluxing stage, one of the outputs is fluxed spectra. They follow the naming convention *OBX*fluxed1000.FIT and *OBX*fluxed1000.FIT, for the 1st and 2nd orders, respectively. They can be downloaded and viewed with fv, as in Figure 3. If for some reason rgsproc wasn't run through that stage, fluxed spectra are easy enough to produce with rgsfluxer, if you have response matrices for both RGS instruments. We'll say that the 1st order RGS2 response matrix is r2_o1_rmf.fits.
For the 1st order, enter rgsfluxer in the Task Name box and click "Change". Then,
- By the pha parameter, use the Browse button to select the spectrum P0153950701R1S001SRSPEC1001.FIT. Another Browse button will appear when a file has been selected; use it to select the other spectrum file, P0153950701R2S002SRSPEC1001.FIT. In a similar way, enter the response matrices r1_o1_rmf.fits and r2_o1_rmf.fits for the parameter rmf.
- Enter the output file name in the parameter file. We will use fluxed_o1.fits.
- Click "Run rgsfluxer".
Fluxed 2nd order spectra are made in a similar way. The spectra are shown in Figure 3. There is good agreement between them, so we can continue.
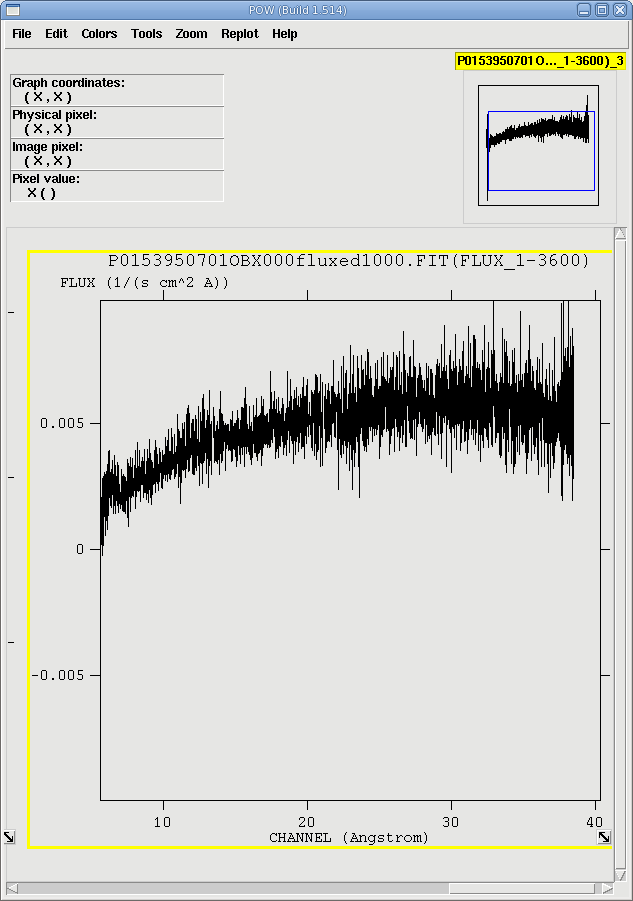
|
Unfortunately, if your spectrum is piled up, there is not a lot you can do to fix it; there is no tried-and-true method like with the EPIC and what there is applies only to certain cases, such as if you assume the 2nd order spectrum is composed entirely of piled up 1st order photons. For more information, see Ness et al. 2007, ApJ, 665, 1334.
Combining Spectra
Spectra from the same order in RGS1 and RGS2 can be safely combined to create a spectrum with higher signal-to-noise if they were reprocessed using rgsproc with spectrumbinning=lambda, which is the default setting. The task we will use to merge source spectra, rgscombine also merges response files and background spectra. When merging response files, be sure that they have the same number of bins. For this example, we assume that RMFs were made for order 1 in both RGS1 and RGS2 with rgsproc, which has a default bin number of 4000.To merge RGS1 and RGS2 spectra, enter rgscombine in the Task Name box and click "Change". Then,
- By the pha parameter, use the Browse button to select the spectrum P0153950701R1S001SRSPEC1001.FIT. Another Browse button will appear when a file has been selected; use it to select the other spectrum file, P0153950701R2S002SRSPEC1001.FIT. In a similar way, enter the response matrices r1_o1_rmf.fits and r2_o1_rmf.fits for the parameter rmf, and the background spectra P0153950701R1S001BGSPEC1001.FIT and P0153950701R2S002BGSPEC1001.FIT for the bkg parameter.
- Enter the output file names in the filepha, filermf, and filebkg parameters; we will use r12_o1_srspec.fits, r12_o1_rmf.fits, and r12_o1_bgspec.fits, respectively.
- In the rmfgrid parameter, enter the number of energy bins, 4000 (this should be the same as the number of energy bins in the input RMFs.)
- Click "Run rgscombine".
If you have any questions concerning XMM-Newton send e-mail to xmmhelp@lists.nasa.gov