THE XMM-NEWTON ABC GUIDE, STREAMLINED
EPIC-PN (TIMING mode), Command Line
Contents
Prepare the Data
Reprocess the Data
Apply Standard Filters
Make a Light Curve
Extract the Source and Background Spectra
Check for Pile Up
My Observation is Piled Up! Now What?
Determine the Spectrum Extraction Areas
Create the Photon Redistribution Matrix (RMF) and Ancillary File (ARF)
Prepare the Data
Please note that the two tasks in this section (cifbuild and odfingest) must be run in the ODF directory. These are the only tasks with that requirement, and after this section, we will work exclusively in our reprocessing directory.Many SAS tasks require calibration information from the Calibration Access Layer (CAL). Relevant files are accessed from the set of Current Calibration File (CCF) data using a CCF Index File (CIF). To set the environment parameter and make the ccf.cif file, type
cd ODF setenv SAS_ODF /full/path/to/ODF/directory/ setenv SAS_ODFPATH /full/path/to/ODF/directory/ cifbuild
To use the updated CIF file in further processing, you will need to reset the environment variable SAS_CCF:
setenv SAS_CCF /full/path/to/ODF/ccf.cif
The task odfingest extends the Observation Data File (ODF) summary file with data extracted from the instrument housekeeping data files and the calibration database. It is only necessary to run it once on any dataset, and will cause problems if it is run a second time. If for some reason odfingest must be rerun, you must first delete the earlier file it produced. This file largely follows the standard XMM naming convention, but has SUM.SAS appended to it. After running odfingest, you will need to reset the environment variable SAS_ODF to its output file. To run odfingest and reset the environment variable, type
odfingest setenv SAS_ODF /full/path/to/ODF/full_name_of_*SUM.SAS
You will likely find it useful to alias these environment variable resets in your login shell (.cshrc, .bashrc, etc.).
Reprocess the Data
To reprocess the data, make a new working directory and call i>epproc.
- epproc
By default, no intermediate files that are generated are kept.
Epproc designates
its output event files with "TimingEvts.ds". In any case, it is
convenient to rename them something easy to type:
- ln -s 1192_0400550201_EPN_S003_TimingEvts.ds pn.fits
Apply Standard Filters
The filtering expression for the PN in TIMING mode is:
(PATTERN <= 4)&&(PI in [200:15000])&&#XMMEA_EP
The first two expressions will select good events with PATTERN in the 0 to 4 range. The PATTERN value is similar the GRADE selection for ASCA data, and is related to the number and pattern of the CCD pixels triggered for a given event. Single pixel events have PATTERN == 0, while double pixel events have PATTERN in [1:4].
The second keyword in the expressions, PI, selects the preferred pulse height of the event. For the PN, this should be between 200 and 15000 eV. This should clean up the image significantly with most of the rest of the obvious contamination due to low pulse height events. Setting the lower PI channel limit somewhat higher (e.g., to 300 or 400 eV) will eliminate much of the rest.
Finally, the #XMMEA_EP filter provides a canned screening set of FLAG values for the event. (The FLAG value provides a bit encoding of various event conditions, e.g., near hot pixels or outside of the field of view.) Setting FLAG == 0 in the selection expression provides the most conservative screening criteria and should always be used when serious spectral analysis is to be done on PN data.
To filter the data, type
evselect table=pn.fits filteredset=pn_filt.fits \
expression='(PATTERN <= 4) && (PI in [200:15000]) && #XMMEA_EP'
where
-
table - input event table
expression - filtering expression
filteredset - output file name
Make a Light Curve
Sometimes, it is necessary to use filters on time in addition to those mentioned above. This is because of soft proton background flaring, which can have count rates of 100 counts/sec or higher across the entire bandpass.To create a light curve, type:
evselect table=pn.fits rateset=pn_ltcrv.fits maketimecolumn=yes timebinsize=50 makeratecolumn=yes
where
-
table - input event table
rateset - name of output light curve file
maketimecolumn - make a time column
timebinsize - time binning (seconds)
makeratecolumn - make a count rate column, otherwise a count column will be created
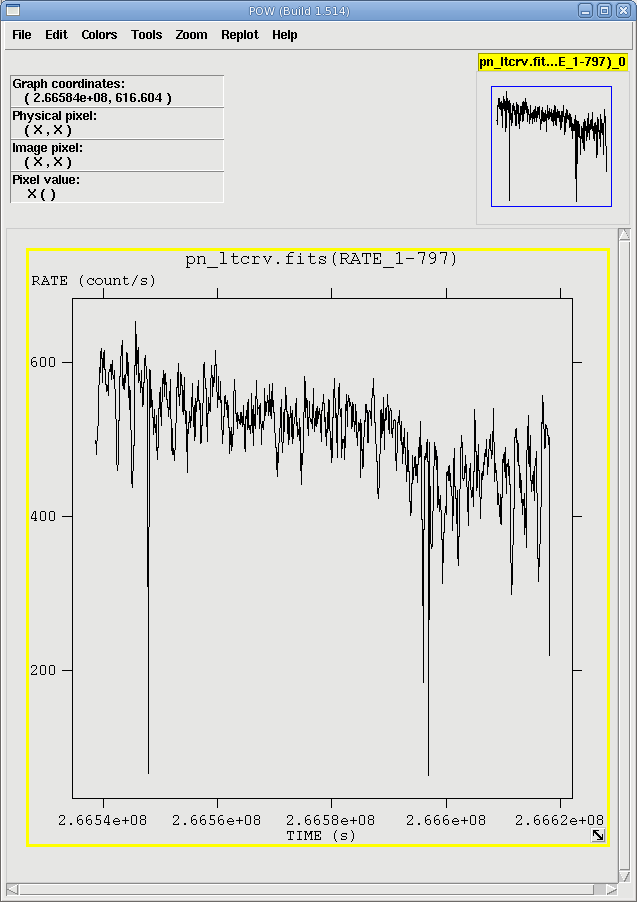
The output file pn_ltcrv.fits can be viewed with fv:
fv pn_ltcrv.fits &
In the pop-up window, the RATE extension will be available in the second row (index 1, as numbering begins with 0). Selecting PLOT from this row will let you choose the column name and axis on which to plot it.
The light curve is shown in Figure 1. No flares are evident, so we will continue to the next section. However, if a dataset does contain flaring, it should be removed in the same way as shown for EPIC IMAGING mode data here.
Extract the Source and Background Spectra
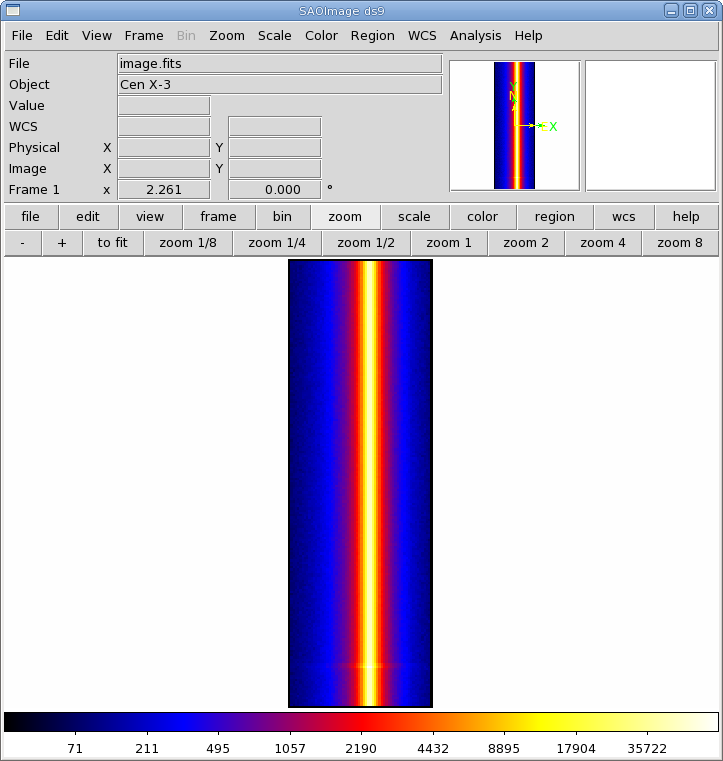
First, we will need to make an image of the filtered event file over the energy range that we are interested in. For this example, we'll say 0.5-15 keV. Since we're using the PN, remember to use the FLAG==0 requirement.
evselect table=pn_filt.fits imagebinning=binSize imageset=image.fits \
xcolumn=RAWX ycolumn=RAWY ximagebinsize=1 yimagebinsize=1 \
expression='(FLAG==0)&&(PI in [500:15000])'
where the keywords are the same as for
applying standard filters, and
-
imagebinning - method of filtering
imageset - name of output image
xcolumn - column with x-coordinates
ycolumn - column with y-coordinates
ximagebinsize - binning factor for x-axis
yimagebinsize - binning factor for y-axis
The image can be displayed with ds9 and is shown in Figure 2. The source is centered on RAWX=37; we will extract this and the 10 pixels on either side of it:
evselect table=pn_filt.fits spectrumset=source_pi_WithBore.fits energycolumn=PI \
spectralbinsize=5 specchannelmin=0 specchannelmax=20479 filteredset=pn_filt_source_WithBore.fits \
expression='(FLAG==0) && (PI in [500:15000]) && (RAWX in [27:47])'
where the keywords are the same as above, and
-
spectrumset - output spectrum name
energycolumn - name of the energy column
spectralbinsize - binning factor for spectrum
specchannelmin - minimum channel number
specchannelmax - maximum channel number
For the background, the extraction area should be as far from the source as possible. However, sources with > 200 ct/s (like our example!) are so bright that they dominate the entire CCD area, and there is no source-free region from which to extract a background. (It goes without saying that this is highly energy-dependent.) In such a case, it may be best not to subtract a background. Users are referred to Ng et al. (2010) for an in-depth discussion. While this observation is too bright to have a good background extraction region, the process is shown below nonetheless for the sake of demonstration:
evselect table=pn_filt.fits spectrumset=bkg_pi.fits energycolumn=PI \
spectralbinsize=5 specchannelmin=0 specchannelmax=20479 filteredset=pn_filt_bkg.fits \
expression='(FLAG==0)&& (PI in [500:15000]) && (RAWX in [3:5])'
Check for Pile Up
Depending on how bright the source is and what modes the EPIC detectors are in, event pile up may be a problem. Pile up occurs when a source is so bright that incoming X-rays strike two neighboring pixels or the same pixel in the CCD more than once in a read-out cycle. In such cases the energies of the two events are in effect added together to form one event. Pile up and how to deal with it are discussed at length here and here, respectively, and users are strongly encouraged to refer to those sections. Briefly, we deal with it in PN TIMING data essentially the same way as in IMAGING data, that is, by using only single pixel events, and/or removing the regions with very high count rates, checking the amount of pile up, and repeating until it is no longer a problem.
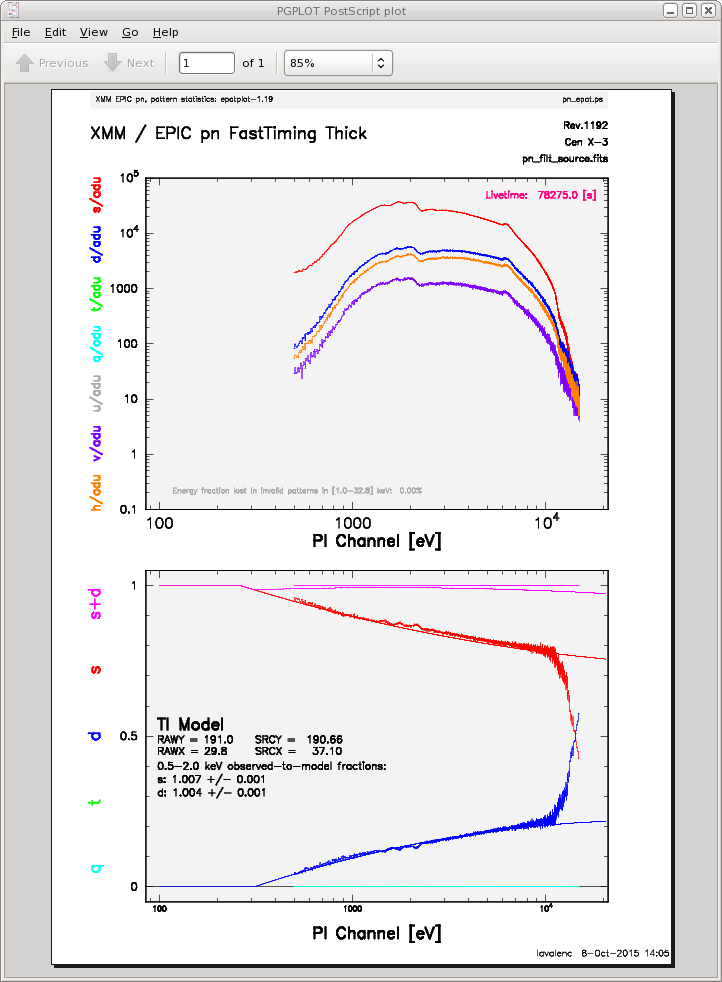
To check for pile up:
epatplot set=pn_filt_source_WithBore.fits plotfile=pn_epat.ps useplotfile=yes \
withbackgroundset=yes backgroundset=pn_filt_bkg.fits
where the keywords are
-
set - the selected region's event file
plotfile - output file name
useplotfile - use "plotfile" as output file name, or default nomenclature?
withbackgroundset - use a background event set for background subtraction
backgroundset - name of background event set
The output is shown in Figure 3. In the lower plot, we can see that there is a large difference between the modeled and observed single and double pattern events, and that the observed-to-model fraction for doubles is over 1.0, indicating that the observation is piled up.
My Observation is Piled Up! Now What?
There are a couple ways to deal with pile up. First, you can use event file filtering procedures to include only single pixel events (PATTERN==0), as these events are less sensitive to pile up than other patterns.
You can also excise areas of high count rates, i.e., the boresight column and several columns to either side of it. (This is analogous to removing the inner-most regions of a source in IMAGING data.) The spectrum can then be re-extracted and you can continue your analysis on the excised event file. As with IMAGING data, it is recommended that you take an iterative approach: remove an inner region, extract a spectrum, check with epatplot, and repeat, each time removing a slightly larger region, until the model and observed pattern distributions agree.
To extract only the columns to either side of the boresight:
evselect table=pn_filt.fits spectrumset=source_pi_NoBore.fits energycolumn=PI \
spectralbinsize=5 specchannelmin=0 specchannelmax=20479 \
expression='(FLAG ==0)&& (PI in [500:15000]) && (RAWX in [27:47]) &&! (RAWX in [29:45])' \
filteredset=pn_filt_source_NoBore.fits
Be aware that if you do this and are using SAS v. 13.x or older, you will need to use a non-standard way to make the ancillary files (ARFs) for your spectrum! This is discussed further in a later section .
Determine the Spectrum Extraction Areas
Now that we are confident that our spectrum is not piled up, we can continue by finding the source and background region areas. (This process is identical to that used for IMAGING data.) This is done with the task backscale, which takes into account any bad pixels or chip gaps, and writes the result into the BACKSCAL keyword of the spectrum table. To find the source and background extraction areas:
backscale spectrumset=source_pi_NoBore.fits badpixlocation=pn_filt.fits
backscale spectrumset=bkg_pi.fits badpixlocation=pn_filt.fits
where
-
spectrumset - spectrum file
badpixlocation - the file containing the bad pixel locations (the event file)
Create the Photon Redistribution Matrix (RMF) and Ancillary File (ARF)
If you are using SAS v. 14 or higher, making the RMF and ARF for PN data in TIMING mode is exactly the same as in IMAGING mode, even if you had to excise piled up areas. This is a change from earlier SAS versions; if you are working with an older SAS, you will need to use the special recipe below to generate the ARF (the method to make a RMF file is the same as shown here.)
To make the RMF,
rmfgen rmfset=source_rmf_NoBore.fits spectrumset=source_pi_NoBore.fits
where
-
rmfset - output file
spectrumset - spectrum file
arfgen arfset=source_arf_NoBore.fits spectrumset=source_pi_NoBore.fits detmaptype=psf \
withrmfset=yes rmfset=source_rmf_NoBore.fits badpixlocation=pn_filt.fits
where
-
arfset - output file
spectrumset - spectrum file
detmaptype - origin of the detector map
withrmfset - use the RMF dataset to define the ARF energy grid?
rmfset - RMF file
badpixlocation - the file containing the bad pixel locations
evselect table=pn_filt.fits spectrumset=source_pi_Excised.fits energycolumn=PI \
spectralbinsize=5 specchannelmin=0 specchannelmax=20479 \
expression='(FLAG ==0)&& (PI in [500:15000]) && (RAWX in [29:45])' \
filteredset=pn_filt_source_Excised.fits
Now we can use the spectra to make the ARFs:
arfgen arfset=source_arf_WithBore.fits spectrumset=source_pi_WithBore.fits detmaptype=psf \
badpixlocation=pn_filt.fits
arfgen arfset=source_arf_Excised.fits spectrumset=source_pi_Excised.fits detmaptype=psf \
badpixlocation=pn_filt.fits
where
-
arfset - output file
spectrumset - spectrum file
detmaptype - origin of the detector map
badpixlocation - the file containing the bad pixel locations
Now we can subtract them:
addarf "source_arf_WithBore.fits source_arf_Excised.fits" "1.0 -1.0" source_arf_NoBore.fits
The spectrum can be fit using HEASoft or CIAO packages, as SAS does not include fitting software.
If you have any questions concerning XMM-Newton send e-mail to xmmhelp@lists.nasa.gov