Next: 7.4 PLT_ENE_BANDS
Up: 7 Reference Guide
Previous: 7.2 FFIT_PREP
FITIT performs forward folding spectrum deconvolution and analysis
via DO_FOLD_ANAL. It takes as input SDB files or SDRs produced by
FFIT_PREP or similar procedures and produces as output best fit models
and pseudo-deconvolved spectra.
FITIT reads in prepared spectra and performs a forward folding
deconvolution analysis, the result of which is a best fit spectrum
model and a pseudo-deconvolved photon spectrum. Log files and plot
files are produced.
Unlike previous (v.7.3 and earlier) versions, FITIT does not expect
the input file to contain a detector sum spectrum in addition to the
individual detector spectra.
Calling Sequence
status = FITIT([FILE = SETUPFILE] [,keywords]
INPUTS
KEYWORDS
*** Initialization and Documentation
- FILE:
- string containing name of namelist setup file. If omitted,
user will be prompted for file name.
- Title:
- Title used for spectrum plot.
- FILEROOT:
- String containing root name for best fit model, deconvolved
spectrum, both SDB and ascii, fit routine diagnostic output,
log and plot files.
Overriden by explicit file name entries (FITMODEL,
PHOTFILE, binfile, fitfile,
logfile, and plotfile).
If not present and explicit entry not present, file will not be produced.
- Logfile:
- String containing name of file containing log information on
run. Defaults to <fileroot>.log. If not set and no FILEROOT,
or if set to 'NONE', no log file is produced.
- Plotfile:
- String containing name of plot file. Defaults to
<fileroot>.ps. If not set and no FILEROOT,
or if set to 'NONE', no plot file is produced.
*** Input Specifications, Selection and Processing
- SDBfile:
- String array(10) containing names of SDB files with
spectra to model. These spectra must either be in
Counts/second, in which case they are interpreted as OSSE
spectra, or
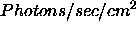 , in which case they are
interpreted as additional data (ie data from other
instruments).
, in which case they are
interpreted as additional data (ie data from other
instruments).
- SDR:
- Optional list of SDRs to be used as input in lieu of SDB file
specified in SETUP. Note that the use of the SDBFILE parameter in
SETUP will override the SDR keyword
- Add_data:
- String array(10) containing file names of files
containing additional data to be fit along with OSSE
data. Data are assumed to be in
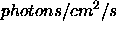 unless units
are encoded otherwise (see below). A unit
response matrix is created to match. Additional data may
overlap OSSE data in energy range, and additional data from
multiple instruments that overlap each other may be
concatenated together into the same file or supplied by the
same function or be supplied in separate files or functions.
All data are concatenated, including additional data supplied
in SDB files specified via the SDBFILE variable (see above).
Data may be presented as a
formatted file readable by the READ_DATAPTS routine.
The order of data is channel edge (MeV), channel width (MeV),
flux integrated over channels (
unless units
are encoded otherwise (see below). A unit
response matrix is created to match. Additional data may
overlap OSSE data in energy range, and additional data from
multiple instruments that overlap each other may be
concatenated together into the same file or supplied by the
same function or be supplied in separate files or functions.
All data are concatenated, including additional data supplied
in SDB files specified via the SDBFILE variable (see above).
Data may be presented as a
formatted file readable by the READ_DATAPTS routine.
The order of data is channel edge (MeV), channel width (MeV),
flux integrated over channels (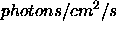 ),
flux uncertainty with one line per spectrum channel.
),
flux uncertainty with one line per spectrum channel.
Although the desired flux units or 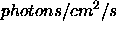 , other units
may be used. In particular, units that are per unit energy
(e.g. /MeV or /keV) may be used. In this case, the comments
of the READ_DATAPTS file must include the phrase
UNITS="*/*MeV*" or UNITS="*/*keV*" or UNITS="@nnnn"
(case insensitive) where the * represents a wild card
and nnnn represents a numeric value of the SPECUNITS variable
from the SDB header in which the /MeV or /keV bits are set.
If this is found, the data are multiplied by the channel widths.
Additional data may also be supplied via a function call. In
this case, ADD_DATA is a string, starting with '@', that
specifies the name of the function to call
(e.g. ADD_DATA='@DATFUNC'). The calling sequence of the
function is:
, other units
may be used. In particular, units that are per unit energy
(e.g. /MeV or /keV) may be used. In this case, the comments
of the READ_DATAPTS file must include the phrase
UNITS="*/*MeV*" or UNITS="*/*keV*" or UNITS="@nnnn"
(case insensitive) where the * represents a wild card
and nnnn represents a numeric value of the SPECUNITS variable
from the SDB header in which the /MeV or /keV bits are set.
If this is found, the data are multiplied by the channel widths.
Additional data may also be supplied via a function call. In
this case, ADD_DATA is a string, starting with '@', that
specifies the name of the function to call
(e.g. ADD_DATA='@DATFUNC'). The calling sequence of the
function is:
DATFUNC(ADDEDG,ADDWID,ADDDAT,ADDSIG,UNITS)
where the first four variables are arrays containing the
channel edges, widths, data and uncertainties and the UNITS
variable is a string specifying the units as described above.
If UNITS is not supplied or does not contain MeV or keV in
the denominator, the units are assumed to be 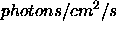 and no modification is undertaken.
and no modification is undertaken.
- Mtrxfile:
- String array (30) containing names of
response matrix files. The matrices
match the data in that, if more than one spectrum is present,
the matrix must have more than one response (each in its own
file). These are assembled into a concatenated matrix in
FITIT. The default is to look for matrices in SDB file aux.
data types.
- TARGNAMES:
- List of target names to use in fit. Default is to use all
targets provided in SDR/RMTRX configuration. Each target name
corresponds to one source spectrum folded through a response
appropriate to that target's position in the collimator for
each count spectrum. Hence, if you desire
a fit to only a subset of all the targets included
in the compound response, TARGNAMES identifies the
desired subset.
- Userscreen:
- String containing name of user screening routine to
select data from SDB files. Calling sequence is:
USERSCREEN,SDRS,GOOD_INDEX
where SDRS is an input array of SDRs with dat,err,edg,wid,crc
populated and GOOD_INDEX is an output index array pointing
to the selected SDRs.
- Detid:
- Integer that specifies which detectors from input SDB files
to use. If input is sum of detectors, if any detectors in
sum match any of criterion detectors, select spectrum
Standard detid bitpick that defaults to 15 (all detectors).
- DSA:
- Integer array(20) that specifies which DSA's to use from input
spectra. Functions if input spectra are SDB files or SDR
keyword SDRs. If all zeroes, use all DSA's (default).
- Scan:
- Float array(20) that specifies which which scan angles to use
from input spectra. Alternative to DSA selection. If both
present, Scan selection is used. Functions if input spectra
are SDB files or SDR keyword SDRs. If all zeroes, use all
scan angles (default). Works by comparing values to targscan
from header, with 0.2 degree tolerance.
- TJD_period:
- Double Precision Array(2,10) containing start and stop
time pairs which specify intervals to include. Format is
day.fraction. Spectra to use must lie entirely within one
of the specified intervals. Default (all zeroes)
is use all intervals.
*** Fitting Specifications
- Modelfile:
- String containing model file name. This file is used for
definition and initialization of the model. If a different
model file is used for fit result output, this initialization
file can be used repeatedly. Defaults to MODELFILE.DAT.
- Fitmodel:
- String containing file name of model file to receive
best fit model. Can be the same as MODELFILE, but this will
overwrite the original file, requiring a new initialization
file for each fit. Alternatively, the output model file
from one fit may be used as the input to another fit.
Defaults to <fileroot>.mdl.
- Photfile:
- String containing file name for SDB file containing
deconvolved photon spectra. Defaults to <fileroot>_PHOT.SDB.
If not set and no FILEROOT,
or if set to 'NONE', or if model has multiple sources (which
implies that the deconvolved photon spectrum is not
meaningful) no SDB file is produced.
- Erange:
- Two element array containing minimum and maximum energies to
fit (in MeV) i.e. [.06,2.] will cause channels from .06 to 2.0
MeV to be fit. Defaults to entire spectrum.
- Nprint:
- Integer that specifies SUPERFIT diagnostic output mode.
See SUPERFIT for details.
- Duper:
- Flag that, when set, activates
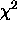 mapping for parameters
with the duper flag set in the model file. Defaults to
cleared (no
mapping for parameters
with the duper flag set in the model file. Defaults to
cleared (no 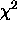 mapping).
mapping).
- Chi2_prob:
- Floating value of the significance level desired
for parameter uncertainties derived via
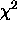 mapping.
Defaults to 0.68.
mapping.
Defaults to 0.68.
- EPS1:
- SUPERFIT iteration termination criterion (see SUPERFIT).
Defaults to 0.0001.
- EPS2:
- SUPERFIT iteration termination criterion (see SUPERFIT).
Defaults to 0.0001.
- Fitfile:
- String containing file name of file to receive SUPERFIT
diagnostic output. Contents depend on value of NPRINT.
Defaults to <fileroot>.dat.
*** Binning Specifications for Plotting
- Binfile:
- String containing file name for ASCII file containing
deconvolved spectrum. Defaults to <fileroot>_PHOT.DAT.
If not set and no FILEROOT,
or if set to 'NONE', no file is produced.
- Plot_bindef:
- String containing name of file or procedure that
defines rebinning for plots.
- Plot_binfrac:
- Flag that, if set allows rebinning with fractional
spectrum channels. Otherwise, integral numbers of energy
channels are added together. Defaults to clear (integral
channels).
- Plot_chanmod:
- String containing code that specifies
alternative means of defining channel summing for plots.
'LOG': PLOT_NCHAN logrithmically spaced channels
'SQRT': PLOT_NCHAN square root channels
'LIN': Add channels in PLOT_NCHAN groups
'NOSUM':No summation
Defaults to 'LOG'. This is overridden by PLOT_BINDEF.
- Plot_nchan:
- Number of summed channels (see above).
*** Display Specifications
- Show_cnts:
- A flag which, if set, results in plots containing count
spectra rather than photon spectra. In either case, the best
fit model is superimposed on the data. Defaults to photon
spectra (i.e. cleared).
- Plotmodels:
- In the case of a model composed of the sum of several
model elements, the number of model elements to plot.
Defaults to plot all elements. Elements are plotted from the
first element to the total requested. For example, if a model
consists of a power law and two gaussian lines, entered in
the model file in that order, setting PLOTMODELS=1 would
result in only the power law being plotted, =2 would result
in only the power law and first line, etc. The only way to
plot the lines without the power law is to enter them first
in the model file (using, for example, BUILDMODEL and MODEL_WRITE), then set PLOTMODELS=2.
- Screenplot:
- Integer flag, set to 1 to produce plots on screen,
0 to produce no plots on screen. Default is produce plots
on screen (1).
- Hardcopy:
- String containing the hardcopy device as used in an IDL
SET_PLOT command (e.g. SET_PLOT,'PS'). Blank => no
hardcopy produced. Defaults to postscript ('PS').
- Plot_erange:
- Two-element array containing minimum and maximum
energies to plot (in MeV) e.g. [.06,2.0] would plot from
.06 to 2.0 MeV. Note that IDL plotting will round the
axes in some cases. Defaults to entire energy range.
- Plot_frange:
- Two-element array containing minimum and maximum fluxes
to plot (in
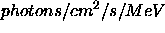 or
or 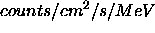 ) e.g.
[1.e-4,2.] would plot from 0.0001 to 2.0
) e.g.
[1.e-4,2.] would plot from 0.0001 to 2.0 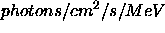 .
Note that IDL plotting will round the
axes in some cases. Defaults to entire flux range.
.
Note that IDL plotting will round the
axes in some cases. Defaults to entire flux range.
- Plot_xtype:
- Plot x-axis linear (0) or log (1). Defaults to log.
- Plot_ytype:
- Plot y-axis linear (0) or log (1). Defaults to log.
- Residuals:
- If set, plot residuals in addition to spectra
and fit results. Defaults to set (residuals plotted).
*** 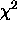 Grid Mapping Specifications
Grid Mapping Specifications
- Mapfile:
- String containing file name for XDR file containing
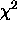 map and related variables. Defaults to <fileroot>_MAP.XDR.
If not set and no FILEROOT, or if set to 'NONE', no file is
produced.
map and related variables. Defaults to <fileroot>_MAP.XDR.
If not set and no FILEROOT, or if set to 'NONE', no file is
produced.
- GENCHIMAP:
- If set, turn on grid-type 1- or 2-Dimensional
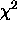 mapping. Default is not set.
mapping. Default is not set.
NOTE: The following parameters define which parameter(s) to map. They
identify the domain, function within the domain (note that many models
may be the sum of several functions) and parameter within the
function.
The output array MAPARRAY, which (for a 2-D map) contains
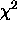 vs parameter-1 and parameter-2 grid point, and the vectors
MAP1 and MAP2, which contain the parameter values for each grid
column and row (respectively), are defined such that performing a
CONTOUR,MAPARRAY,MAP1,MAP2 will align parameter-1 with the x-axis of
the plot and parameter-2 with the y-axis of the plot.
vs parameter-1 and parameter-2 grid point, and the vectors
MAP1 and MAP2, which contain the parameter values for each grid
column and row (respectively), are defined such that performing a
CONTOUR,MAPARRAY,MAP1,MAP2 will align parameter-1 with the x-axis of
the plot and parameter-2 with the y-axis of the plot.
- MAPPARDOMAIN1:
- "domain" or source spectrum containing parameter-1 for
two dimensional
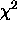 mapping
(or the only parameter in the case of 1-D mapping). This
is a string containing a target name. The target name is
converted into a target id number, which is compared to the
domain indices in the matrix/model structure. Remember that
each domain is one source spectrum and hence is identified by
a single target id. If this variable is null, then the
first domain will be used regardless of target id.
String array of 30 elements, each of which defines the
domain for a 1D map of one parameter. For 2-D map, only
first element is used.
mapping
(or the only parameter in the case of 1-D mapping). This
is a string containing a target name. The target name is
converted into a target id number, which is compared to the
domain indices in the matrix/model structure. Remember that
each domain is one source spectrum and hence is identified by
a single target id. If this variable is null, then the
first domain will be used regardless of target id.
String array of 30 elements, each of which defines the
domain for a 1D map of one parameter. For 2-D map, only
first element is used.
- MAPPARDOMAIN2:
- "domain" or source spectrum containing parameter-2
for two dimensional
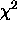 mapping. This
is a target name string (see above).
mapping. This
is a target name string (see above).
- MAPPARFUNC1:
- Within the domain defined by MAPPARDOMAIN1, this
is a string containing the call name of the function
(e.g. Pwrlaw for a power law) containing the desired
parameter. String array of 30 elements, each
of which defines the function for a 1-D map of one parameter.
For 2-D map, only first element is used.
- MAPPARFUNC2:
- Within the domain defined by MAPPARDOMAIN2, this
is a string containing the call name of the function
containing the desired parameter.
- MAPPARFUNC1OCC:
- Since some models may have multiple occurances of
a particular function within a single domain (e.g. multiple
Gaussian line functions), this is an integer that identifies
which occurance of the function specified in MAPPARFUNC1
contains the desired parameter. 1 is the first occurance,
etc. Defaults to 1. Integer array of 30 elements, each
of which defines the occurance number for a 1-D map
of one parameter. For 2-D map, only first element is used.
- MAPPARFUNC2OCC:
- Since some models may have multiple occurances of
a particular function within a single domain (e.g. multiple
Gaussian line functions), this is an integer that identifies
which occurance of the function specified in MAPPARFUNC2
contains the desired parameter. 1 is the first occurance,
etc. Defaults to 1.
- MAPPAR1:
- Parameter sequence number within the specified function of
the desired first parameter. Integer array of 30 elements, each
of which defines the parameter number for a 1-D map
of one parameter. For 2-D map, only first element is used.
- MAPPAR2:
- Parameter sequence number within the specified function of
the desired parameter.
- MAPPAR1INC:
- Increment in parameter-1 between grid values.
Floating array of 30 elements, each
of which defines the parameter increment for a 1-D map
of one parameter. For 2-D map, only first element is used.
- MAPPAR2INC:
- Increment in parameter-2 between grid values.
- NMAPPAR1:
- Number of grid points to each side of the best fit point
for parameter-1 (i.e. 2*NMAPPAR1 +1 points). Integer array of 30 elements,
each of which defines the number of parameters for a 1-D map of one parameter.
For 2-D map, only first element is used.
- NMAPPAR2:
- Number of grid points to each side of the best fit point
for parameter-2 (i.e. 2*NMAPPAR2 +1 points).
Outputs
Outputs include the following files:
- LOGFILE:
- file containing history of run
- PLOTFILE:
- file containing PostScript plots of deconvolved spectra
- FITFILE:
- file containing SUPERFIT output as specified by NPRINT
- FITMODEL:
- modelfile containing output best fit model
- PHOTFILE:
- SDB file containing deconvolved spectra
- BINFILE:
- ASCII file containing deconvolved spectra binned as specified
Next: 7.4 PLT_ENE_BANDS
Up: 7 Reference Guide
Previous: 7.2 FFIT_PREP
![]() , other units
may be used. In particular, units that are per unit energy
(e.g. /MeV or /keV) may be used. In this case, the comments
of the READ_DATAPTS file must include the phrase
UNITS="*/*MeV*" or UNITS="*/*keV*" or UNITS="@nnnn"
(case insensitive) where the * represents a wild card
and nnnn represents a numeric value of the SPECUNITS variable
from the SDB header in which the /MeV or /keV bits are set.
If this is found, the data are multiplied by the channel widths.
Additional data may also be supplied via a function call. In
this case, ADD_DATA is a string, starting with '@', that
specifies the name of the function to call
(e.g. ADD_DATA='@DATFUNC'). The calling sequence of the
function is:
, other units
may be used. In particular, units that are per unit energy
(e.g. /MeV or /keV) may be used. In this case, the comments
of the READ_DATAPTS file must include the phrase
UNITS="*/*MeV*" or UNITS="*/*keV*" or UNITS="@nnnn"
(case insensitive) where the * represents a wild card
and nnnn represents a numeric value of the SPECUNITS variable
from the SDB header in which the /MeV or /keV bits are set.
If this is found, the data are multiplied by the channel widths.
Additional data may also be supplied via a function call. In
this case, ADD_DATA is a string, starting with '@', that
specifies the name of the function to call
(e.g. ADD_DATA='@DATFUNC'). The calling sequence of the
function is: ![]() and no modification is undertaken.
and no modification is undertaken.