While a variety of analysis packages can be used for the following steps, the SAS was designed for the basic reduction and analysis of XMM-Newton data (extraction of spatial, spectral, and temporal data); therefore, it will be used here for demonstration purposes.
NOTE: For PN observations with very bright sources, out-of-time
events can provide a serious contamination of the image. Out-of-time
events occur because the read-out period for the CCDs can be up to
![]() % of the frame time. Since events that occur during the
read-out period can't be distinguished from others events, they are
included in the event files but have invalid locations. For
observations with bright sources, this can cause bright stripes
in the image along the CCD read-out direction.
% of the frame time. Since events that occur during the
read-out period can't be distinguished from others events, they are
included in the event files but have invalid locations. For
observations with bright sources, this can cause bright stripes
in the image along the CCD read-out direction.
At this point, it is assumed that you have downloaded the data from the HEASARC archive onto a Hera server, standard or anonymous Hera is running (see §4.2), and you have prepared the data for processing (see §6). Throughout this chapter, as in §6, we will use the Lockman Hole dataset with ObsID 0123700101, though any dataset will suffice.
If a dataset is more than a year old, it was probably processed with older versions of CCF and SAS prior to archiving, so the pipeline should be rerun to generate event files with the latest calibrations. The MOS has two pipeline tasks, emchain and emproc, while the PN has one, epproc for the PN. The two MOS tasks produce the same output, so which one to use is entirely a matter of the user's personal preference.
Verify that the working directory PROC is highlighted in the GUI. In the
new Command Window you made at the end of §6, run the task(s):
or
and
If the dataset has more than one exposure, a specific exposure can be accessed using the exposure parameter, e.g.:
where n is the exposure number. To create an out-of-time event file for your PN data, add the parameter withoutoftime to your epproc invocation:
By default, none of these tasks keep any intermediate files they generate. Emchain
maintains the naming convention described in §5.3.3.
Emproc and epproc designate their output event files with ``Evts.ds'';
``*ImagingEvts.ds'', ``*TimingEvts.ds'', and ``*BurstEvts.ds'' denote the imaging mode, timing,
and burst mode event lists, respectively. In either case, you may want to name the new files
something easy to type. Right-clicking on a file will give you the option to rename it.
Once the new event files have been obtained, the analysis techniques described below can be used. We will refer to the new event files as mos1.fits, mos2.fits, and pn.fits.
To create an image in sky coordinates, type
where
The resultant image is written to the file image.fits. It can be viewed with
POW, or downloaded to your local machine and viewed with ds9;
see Figure 7.1.
To create a light curve, type
where
The output file mos1_ltcrv.fits can be viewed with POW; see Fig. 7.2.
The filtering expressions for the MOS and PN are:
and
If the PN data is timed, then the PATTERN parameter should be set to 4:
The first two expressions will select good events with PATTERN in the 0 to 12 range, and the last will select events with PATTERN between 0 and 4. The PATTERN value is similar the GRADE selection for ASCA data, and is related to the number and pattern of the CCD pixels triggered for a given event.The PATTERN assignments are: single pixel events: PATTERN == 0, double pixel events: PATTERN in [1:4], triple and quadruple events: PATTERN in [5:12].
The second keyword in the expressions, PI, selects the preferred pulse height of the event; for the MOS, this should be between 200 and 12000 eV. For the PN, this should be between 200 and 15000 eV. This should clean up the image significantly with most of the rest of the obvious contamination due to low pulse height events. Setting the lower PI channel limit somewhat higher (e.g., to 300 eV) will eliminate much of the rest.
Finally, the #XMMEA_EM (#XMMEA_EP for the PN) filter provides a canned screening set of FLAG values for the event. (The FLAG value provides a bit encoding of various event conditions, e.g., near hot pixels or outside of the field of view.) Setting FLAG == 0 in the selection expression provides the most conservative screening criteria and should always be used when serious spectral analysis is to be done on the PN.
It is a good idea to keep the output filtered event files and use them in your
analyses, as opposed to re-filtering the original file with every task. This will
save much time and computer memory. As an example, the Lockman Hole data's original
event file is 48.4 Mb; the fully filtered list (that is, filtered spatially, temporally,
and spectrally) is only 4.0Mb!
To filter the data, type
where
Sometimes, it is necessary to use filters on time in addition to those mentioned above. This is because of soft proton background flaring, which can have count rates of 100 counts/sec or higher.
It should be noted that the amount of flaring that needs to be removed depends in part on the object observed; a faint, extended object will be more affected than a very bright X-ray source.
There are two ways to filter on time: with an explicit reference to the TIME or RATE parameters in the filtering expression, or by creating a secondary Good Time Interval (GTI) file with the task tabgtigen. Both procedures are described below. For the example data, we will filter by time, though you can just as easily filter by rate.
To explicitly define the TIME or RATE parameters, make a light curve
and display it, as demonstrated in §7.2.2 and plotted in
Figure 7.2. There is a very large flare toward the end of the
observation, so the syntax for the time selection is (TIME ![]() 7.32273e7).
However, there is also a small flare within an otherwise good interval. A slightly
more comlicated expression to remove it would be:
(TIME
7.32273e7).
However, there is also a small flare within an otherwise good interval. A slightly
more comlicated expression to remove it would be:
(TIME ![]() 7.32273e7)&&!(TIME IN [7.32219e7:7.32238e7]). The syntax
&&(TIME
7.32273e7)&&!(TIME IN [7.32219e7:7.32238e7]). The syntax
&&(TIME ![]() 7.32273e7) includes only events with times less than 7.32273e7, and
the ``!'' symbol stands for the logical ``not'', so use
&&!(TIME in [7.32219e7:7.32238e7])
to exclude events in the time interval 7.32219e7 to 7.32238e7.
7.32273e7) includes only events with times less than 7.32273e7, and
the ``!'' symbol stands for the logical ``not'', so use
&&!(TIME in [7.32219e7:7.32238e7])
to exclude events in the time interval 7.32219e7 to 7.32238e7.
If combined with the standard filtering expression (see §7.2.3), the full filtering expression would then be:
This expression can then be used to filter the original event file, as shown in §7.2.3, or only the times can be used to filter the file that has already had the standard filters applied:
where the keywords are as described in §7.2.3.
To filter on time using a secondary GTI file, make the file by using the same time filtering parameters as determined above and the tabgtigen task,
where
and apply the new GTI file with the evselect task:
where
The task edetect_chain is a metatask that does nearly all the work involved
with EPIC source detection. It can take as input arbitrary combinations of images from
different energy bands and different EPIC instruments, with up to three instruments
and 5 energy bands. However, users should be aware that that the binning and WCS
keywords in all images must be identical.
Edetect_chain is comprised of seven straightforward tasks that can
also be run by hand. Edetect_chain requires input files to be generated
and prepared using the tasks atthkgen and evselect; the task
emosaic, while not necessary for source detection, does provide a nice
mosaicked image for display purposes. Fortunately, these are all quick and
straightforward.
In the example below, source detection on images is done in
two bands, 500 - 1000 eV and 4500 - 12000 eV, which correspond to bands 2 and 5 of the
3XMM Catalogue,
for all three detectors. The source count rates are converted into fluxes
through the energy converstion factors (ECFs) for each detector and energy
band. The ECFs depend on the pattern selection and filter used during the observation
and are given in units of ![]() cts cm
cts cm![]() /erg. Interested users can find all
of the bands listed in
Table 1
of the Catalogue. The ECFs are in
Table 8
of the Catalogue.)
/erg. Interested users can find all
of the bands listed in
Table 1
of the Catalogue. The ECFs are in
Table 8
of the Catalogue.)
The example uses the filtered event files from all three cameras, which can be
produced in §7.2.4.
First, make the attitude file:
where
Next, make the band 2 and band 5 images with evselect. We'll start with the band 2 image in the MOS1.
and
The procedure is similar for the MOS2 and PN. Note that, because we are using these
images for the specific purpose of source detection, the image binning that we would
normally use for MOS (22, corresponding to 1.1 arcsec) must be adjusted to match that
of PN (82, corresponding to 4.1 arcsec). For the band 5 images, set the
``Selection Expression'' text to (FLAG == 0)&&(PI in [4500:12000]).
We will assume the output images are named mos1-b2.fits, mos2-b2.fits, pn-b2.fits,
mos1-b5.fits, mos2-b5.fits, and pn-b5.fits.
Now we can run edetect_chain:
where
Throughout the following, please keep in mind that some parameters are instrument-dependent. The parameter specchannelmax should be set to 11999 for the MOS, or 20479 for the PN. Also, for the PN, the most stringent filters, (FLAG==0)&&(PATTERN<=4), must be included in the expression to get a high-quality spectrum.
For the MOS, the standard filters should be appropriate for many cases, though there are some instances where tightening the selection requirements might be needed. For example, if obtaining the best-possible spectral resolution is critical to your work, and the corresponding loss of counts is not important, only the single pixel events should be selected (PATTERN==0). If your observation is of a bright source, you again might want to select only the single pixel events to mitigate pile up (see §7.2.8 and §7.2.9 for a more detailed discussion).
First, make an image of the filtered file, mos1_filt_time.fits, as described in §7.2.1). Download it and display it with ds9, then click on an object whose spectrum you wish to extract. Adjust the extraction region until you are satisfied with it. For this example, we will choose the source at (26165.75, 22816.25) and set the extraction radius to 400.
To extract the source spectrum, type
where
The spectrum, in counts per channel, can be viewed with fv.
When extracting the background spectrum, follow the same procedures, but change the
extraction area. For example, make an annulus around the source; this can be done
using two circles, each defining the inner and outer edges of the annulus, then
change the filtering expression (and output file name) as necessary.
To extract the background spectrum, type
where the keywords are as described above.
The source and background region areas can now be found. This is done with the task
backscale, which takes into account any bad pixels or chip gaps, and writes
the result into the BACKSCAL keyword of the spectrum table.
To find the source and background extraction areas, type
and
Depending on how bright the source is and what modes the EPIC detectors are in, event pile up may be a problem. Pile up occurs when a source is so bright that incoming X-rays strike two neighboring pixels or the same pixel in the CCD more than once in a read-out cycle. In such cases the energies of the two events are in effect added together to form one event. If this happens sufficiently often, 1) the spectrum will appear to be harder than it actually is, and 2) the count rate will be underestimated, since multiple events will be undercounted. To check whether pile up may be a problem, use the SAS task epatplot. (Heavily piled sources will be immediately obvious, as they will have a ``hole'' in the center.) Note that this procedure requires as input the event files created when the spectrum was made.
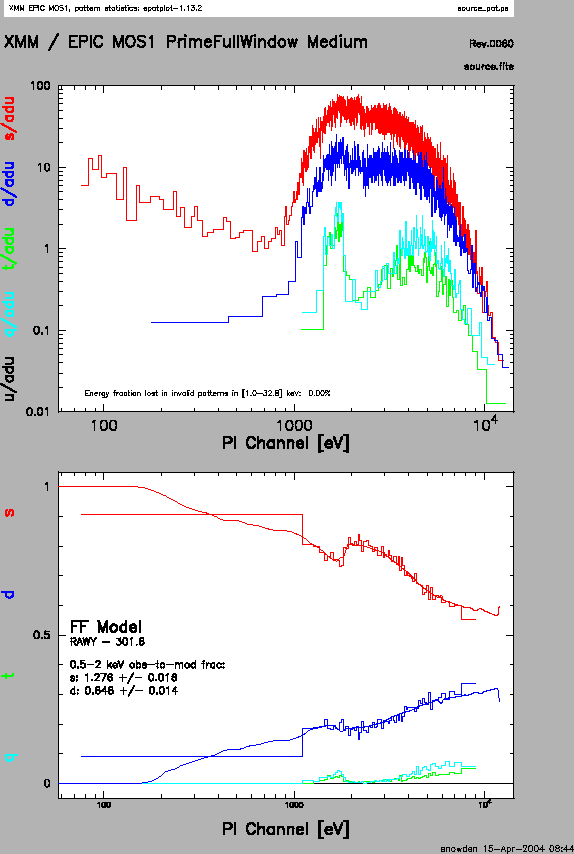
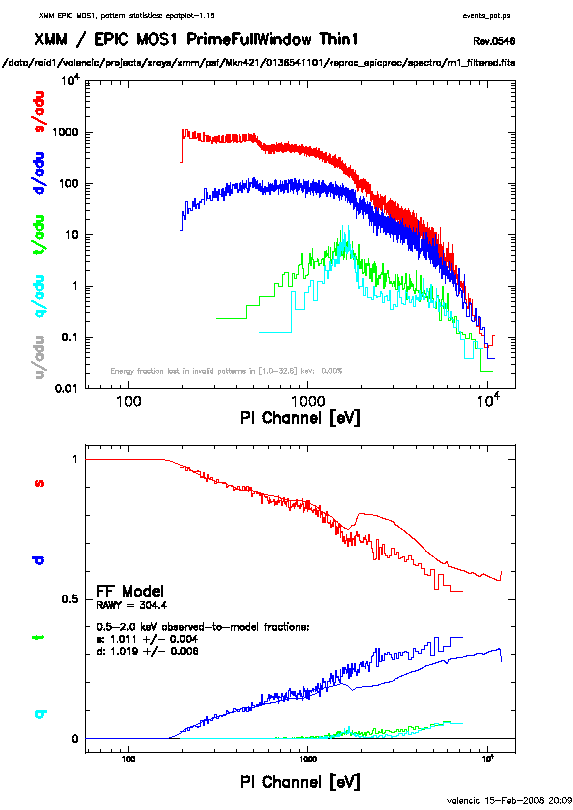
The output of epatplot is a postscript file, which may be viewed with viewers such as gv, containing two graphs describing the distribution of counts as a function of PI channel; see Figure 7.3.
A few words about interpretting the plots are in order. The top is the distribution of counts versus PI channel for each pattern class (single, double, triple, quadruple), and the bottom is the expected pattern distribution (smooth lines) plotted over the observed distribution (histogram). If the lower plot shows the model distributions for single and double events diverging significantly from the observed distributions, then the source is piled up.
The source used in our Lockman Hole example is too faint to provide reasonable
statistics for epatplot and is far from being affected by pile up.
In contrast, plots from two different observations are shown in
Figure 7.3 and 7.4. In Figure 7.3,
the source is bright enough to provide statistics (and a good fit) at energies above
about 1.5 keV. Figure 7.4 shows a plot of a very bright source
which is strongly affected by pileup. Note the severe divergence
between the model and the observed pattern distribution.
To check for pile up, type
The postscript file can be copied to the user's local machine and viewed there.
 |
 |
There are a few ways to deal with pile up. First, using the region selection and event file filtering procedures demonstrated in earlier sections, you can excise the inner-most regions of a source (as they are the most heavily piled up), re-extract the spectrum, and continue your analysis on the excised event file. For this procedure, it is recommended that you take an iterative approach: remove an inner region, extract a spectrum, check with epatplot, and repeat, each time removing a slightly larger region, until the model and observed distribution functions agree.
You can also use the event file filtering procedures to include only single pixel events (PATTERN==0), as these events are less sensitive to pile up than other patterns.
In order to do spectral analysis, it is necessary to find the instrument's
repsonse as a function of energy and PI channel. This is done by reformating
the detector response and energy bounds information and correcting for
instrumental effects, and writing the result to the Redistribution Matrix
File (RMF). The following assumes that an appropriate source spectrum, named
mos1_source_pi.fits, has been extracted as in §7.2.6.
To make the RMF, type
where
Now use the RMF, spectrum, and event file to make the ancillary file.
To make the ARF, type
where
At this point, the spectrum is ready to be analyzed, so we can prepare the spectrum for fitting.
With the source and background spectra now extracted and the RMF and ARF created, we will do some simple spectral fitting. SAS does not include fitting software, so HEASoft packages will be used, and all fitting tasks will be called from the Command Window.
Nearly all spectra will need to be binned for statistical purposes. The procedure grppha, located in the HEASARC folder in the Available Tools window, provides an excellent mechanism to do just that.
The following commands not only group the source spectrum for Xspec but also associate the appropriate background and response files for the source.
and edit the parameters and file names as appropriate:
Please enter PHA filename[] mos1_source_pi.fits ! input spectrum file name
Please enter output filename[] mos1_grp.fits ! output grouped spectrum
GRPPHA[] chkey BACKFILE mos1_bkg_pi.fits ! include the background spectrum
GRPPHA[] chkey RESPFILE mos1_rmf.fits ! include the RMF
GRPPHA[] chkey ANCRFILE mos1_arf.fits ! include the ARF
GRPPHA[] group min 25 ! group the data by 25 counts/bin
GRPPHA[] exit
Upon exiting, the output file mos1_grp.fits will appear in your working directory.
Next, use Xspec to fit the spectrum by typing:
A POW window will pop up and display the spectrum later on. Edit the parameters and file names as appropriate:
XSPEC> data mos1_grp.fits ! input data
XSPEC> ignore 0.0-0.2,6.6-** ! ignore unusable energy ranges, in keV
! set a range appropriate for the data
XSPEC> model wabs(pow+pow) ! set spectral model to two absorbed power laws
1:wabs:nH> 0.01 ! set model absorption column density to 1.e20
2:powerlaw:PhoIndex> 2.0 ! set the first model power law index to -2.0
3:powerlaw:norm> ! use the default model normalization
4:powerlaw:PhoIndex> 1.0 ! set the second model power law index to -1.0
5:powerlaw:norm> ! use the default model normalization
renorm ! renormalize the model spectrum
XSPEC> fit ! fit the model to the data
XSPEC> setplot energy ! plot energy along the X axis
XSPEC> plot ldata ratio ! plot two panels with the log of the data and
! the data/model ratio values along the Y axes
XSPEC> exit ! exit Xspec
Figure 7.5 shows the fit to the spectrum.
This section will demonstrate some basic timing analysis of EPIC image-mode data using the Xronos analysis package. For this exercise, the central source from the observation of G21.5-09 (Obs ID 0122700101) is used. These examples assume that the source's lightcurve has been made as in §7.2.2, but with timebinsize set to 1 and makeratecolumn set to no; the name of this file assumed to be source_ltcrv.fits.
For the aficionado, the task barycen can be used for the barycentric correction of the source event arrival times.
The Xronos tools can be access through Hera by clicking on XRONOS
in the Available Tools panel.
To make a binned lightcurve, type:
where
The output can be viewed with fv by right-clicking on the filename and selecting the
``Edit/Display File'' option. Output is shown in Figure 7.6.
To calculate power spectrum density, type
where
The output can be viewed with fv by right-clicking on the filename and selecting the
``Edit/Display File'' option. Output is shown in Figure 7.7.
To search for periodicities in the time series, type:
where
To calculate the autocorrelation for a time series, type:
where
To calculate statistical quantities for a time series, type
where
The output will be written in the Command Window.