Table of Contents
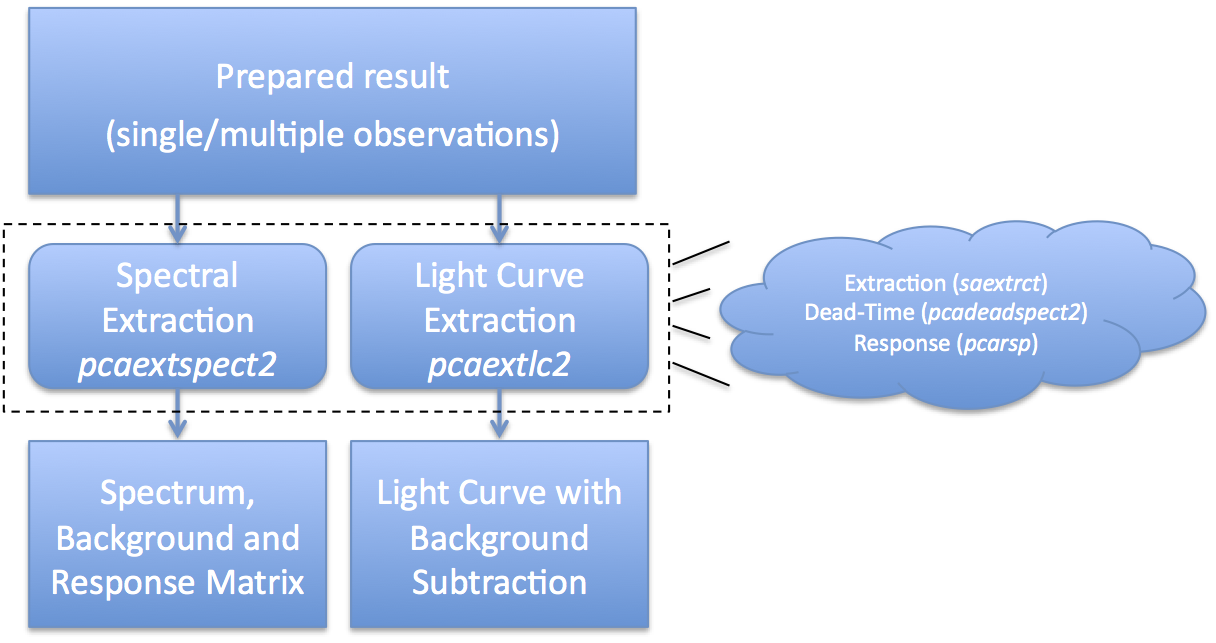
IntroductionAt this point, you are at the step where you extract scientific products. The products we will discuss here are Standard2 spectra, obtained with the task 'pcaextspect2', and light curves, obtained with the task 'pcaextlc2'. These tasks are very similar, and differ only in the details peculiar to spectra or light curves. Figure 1 gives an overview of the extraction process. What Figure 1 shows is that you start with a set of prepared results, either from a single observation or the result of merging multiple observations. You run the task 'pcaextspect2' to obtain spectra and associated metadata, also known as PHA and RSP files; and 'pcaextlc2' to obtain light curves, also known as LC files. Internally, these extractors use the well-known standard tools like 'saextrct' for extraction and 'pcarsp' for response generation. pcaextspect2 extracts a spectrum from PCA data stored in the Standard2 format. This task performs spectrum extraction for both source and background, dead-time correction, and optionally calculates a PCA response matrix which corresponds to the observation. pcaextspect2 is a "convenience" script which makes it easier and less error-prone to extract a spectrum for most users. Extracting Spectra with 'pcaextspect2'For spectra, it is expected that you will extract both a source spectrum and a background spectrum. Every RXTE PCA observation has an associated background, and you must include background in your subsequent analysis. By default, both spectra are dead-time corrected. Also by default, each spectrum should have an associated PCA response matrix which provides information about the spectral sensitivity during your particular set of observations. You should not use a generic response matrix. The task 'pcaextspect2' can be run interactively from the command line. Here is an example. csh> pcaextspect2 > Input Standard2 file name or @file-of-filenames: [] @93067-all-result/FP_dtstd2.lis > Input background file name or @file-of-filenames: [] @93067-all-result/FP_dtbkg2.lis > Output dead-time corrected source spectrum: [] 93067-all_src.pha > Output dead-time corrected background spectrum: [] 93067-all_bkg.pha > Input GTI file to be AND'd with INFILE:[] 93067-all-basic.gti > Comma-separated list of PCU detectors to accmulate (0,1,2,3,4): [] ALL > Comma-separated list of PCU layers to accmulate (1,2,3): [] ALL > Name of output response matrix file (or NONE): [] 93067-all.rsp > Name of XTE filter file (or NONE): [] @93067-all-result/FP_xtefilt.lis This seems like a large number of parameters, but it is just enough to specify all of the relevant inputs and outputs. Here is a quick description of each parameter as entered:
The same effect can be achieved non-interactively with the following command. pcaextspect2 \
src_infile=@93067-all-result/FP_dtstd2.lis \
bkg_infile=@93067-all-result/FP_dtbkg2.lis \
src_phafile=93067-all_src.pha bkg_phafile=93067-all_bkg.pha \
gtiandfile=93067-all-basic.gti \
filtfile=@93067-all-result/FP_xtefilt.lis \
respfile=93067-all.rsp \
pculist=ALL layerlist=ALL
Note that it is safe to specify pculist=ALL even if some PCUs are off or a high voltage breakdown occurred. The task is smart enough to tally only the live time of the enabled good detectors, and exclude the bad detectors. Details of Spectral ExtractionA key benefit is that users do not need special knowledge of when detectors become "dead" for various reasons. Instead, the user can request all of the detectors they are interested in, and 'pcaextspect2' will produce a spectrum which contains the subset of those detectors which have good live time. For example, users can request all of the detectors all of the time (pculist="0,1,2,3,4"), and pcaextspect2 will automatically de-select the detectors which are disabled, or having a breakdown, etc. This functionality relies on the "zero_bad" capabilities of the "pcadeadcalc2" task. Advanced users can still use the more basic tools for fine-grained control instead of this task. Because extraction and dead-time correction require the user to perform several careful operations, the use of this tool is recommended to make sure that the operations are done consistently. Generally, the PCA team and the RXTE GOF recomend to not change the default for the advance parameters of this tool without specific need. The input to this tool is a set of PCA Standard2 files. To gain full benefit of this tool, the user MUST provide dead-time calculated version of the Standard2 file. For beginning users, the output files of the 'pcaprepfile2' or 'pcaprepobsid' tasks are sufficient. For advanced users, the equivalent output of 'pcadeadcalc2' is acceptable. The user must provide both the source and background Standard2 files. Beginning users can use the output of 'pcaprepfile2' / 'pcaprepobsid'; advanced users may use the output of 'pcabackest' if desired. The output of the task is a source spectrum file (.pha file) and a background spectrum file for the same time interval. These files are dead-time corrected, and contain only counts and exposure from live detectors, as described in the help file for the task 'pcadeadcalc2'. The output will be the total counts spectrum of all live detectors, with an exposure that is the sum of the exposures for the individual PCU detectors. Thus, the rate spectrum appearing in software like XSPEC will have units of "counts per detector" ("counts per PCU"). The task can also optionally calculate a response matrix. This matrix is tailored for the specific observation and weighted appropriately. In this case, users must specify the filter file using the 'filtfile' parameter. Recommendations for PCU and Layer SelectionsThe subsection has a few recommendations about which PCU detectors and/or layers to choose using the 'pculist' and 'layerlist' parameters. This selection can make a difference in some cases. For faint sources, fainter than about 20 mCrab = 5 x 10-10 erg s-1 cm-2 = 40 ct/s/PCU, most of the photons will be absorbed in the top layer, and few source photons will survive to layer 2 or layer 3. Layers 2 and 3 will likely be background dominated and will contribute more noise than signal to your results. Therefore, the PCA team typically recommends that for targets <20 mCrab, use only layer 1 ("layerlist=1"). Generally speaking, users are recommended to use all available PCU detectors (pculist=ALL). The software will automatically disregard detectors that are dead in any way (see the task 'pcadeadcalc2'), so this is a safe setting even if not all PCUs are on. The intercalibration between PCUs at end of mission is quite good (~0.5% systematic residuals for a Crab-like spectrum), so there is little harm in including counts from different PCUs. Faint sources will benefit more from counts gained (Poisson counting errors are larger than systematic background or response errors of ~1%), so for these targets use pculist=ALL. For bright targets this is less important and it's acceptable to just use a single PCU. By default, data from PCUs 0 and 1 are excluded by the software discussed here, after those detectors lost their propane layer. The PCA team does not normally recommend analyzing this data without great care. Manually Calculating a Response Matrix LaterIf users wish to calculate their own response matrix at a later time, outside of pcaextspect2, they can use 'pcarsp' task. Because of the complexity of selecting the proper detectors and layers, the RXTE GOF and PCA teams recommend to allow this task to compute the response matrix, but it can be done "by hand." The important parameters for calling pcarsp are, -l (choose same layer list as used to extract spectrum) -p (choose same detector list as used to extract spectrum) -w INFILE (applies appropriate PCU weights for observation) The output spectrum produced by this task contains PCU weighting keywords than can be used automatically by selecting the "-w" parameter. More information about creating PCA response matrices can be found at the PCA response generation page. Combining Spectra from Different ExtractionsIf the user wishes to combine spectra produced by pcaextspect2 in some form of downstream processing, then these instructions may be of help. Users should combine the spectra with the following general settings:
For the task 'mathpha' the settings should be
Of course, this operation needs to be performed separately for both the source and background files. To combine the responses, the user should use the 'addrmf' task and an input file with weights. The weights should be calculated according to the relative exposure. For example, if spectrum1.pha has 1000 seconds of exposure and spectrum2.pha has 500 seconds of exposure then the weighting file should look like this: spectrum1.rsp 0.6667 spectrum2.rsp 0.3333 Note how 0.6667 equals (1000/1500) and 0.3333 equals (500/1500), where 1500 seconds is the total exposure of both. By definition, the weights in the file must sum to 1.0. Using more than one GTIIf you have more than one good time interval file (GTI), then it is easy to combine them. Make as many different filterings as you need using 'maketime' or some other filtering program. When you run the task 'pcaextspect2' the 'gtiandfile' parameter can accept a comma-separated list of GTI file names, which will be logically ANDed together. In other words, the union of the GTIs that you specify will be used to filter the data. You can also use a the @filename.lis convention to list your GTIs in another filename.lis file. Extracting Light Curves with 'pcaextlc2'pcaextlc2 extracts a light curve from PCA data stored in the Standard2 format. This task performs light curve extraction for both source and background, and optionally performs both background subtraction and/or dead-time correction. pcaextlc2 is a "convenience" script which makes it easier and less error-prone to extract a light curve for most users. A key benefit is that users do not need special knowledge of when detectors become "dead" for various reasons. Instead, the user can request all of the detectors they are interested in, and 'pcaextlc2' will produce a light curve which contains the subset of those detectors which have good live time, and is represents the average rate per detector. For light curves, it is expected that you will be interested in both the source rate and background rate. By default, extracted light curves are background-subtracted and dead-time corrected. The task 'pcaextspect2' can be run interactively from the command line. Here is an example. csh> pcaextlc2 > Input Standard2 file name or @file-of-filenames: [] @93067-all-result/FP_dtstd2.lis > Input background file name or @file-of-filenames: [] @93067-all-result/FP_dtbkg2.lis > Output light curve file: [] 93067-all.lc > Input GTI file to be AND'd with INFILE:[] 93067-all-basic.gti > Comma-separated list of PCU detectors to accmulate (0,1,2,3,4): [] ALL > Comma-separated list of PCU layers to accmulate (1,2,3): [] ALL > Light curve time bin size (multiple of 16 sec): (0:) [] 16 Here is a quick description of each parameter as entered:
The same effect can be achieved non-interactively with the following command. pcaextlc2 \
src_infile=@93067-all-result/FP_dtstd2.lis \
bkg_infile=@93067-all-result/FP_dtbkg2.lis \
outfile=93067-all.lc \
gtiandfile=93067-all-basic.gti \
pculist=ALL layerlist=ALL binsz=16
For Standard2 data, the time bin size must be a multiple of 16 seconds. Selecting a Specific Energy Range (also Hardness Ratios)By default, the task 'pcaextlc2' will choose the entire channel (energy) range. This is rarely what is desired. Usually the user is interested in a particular energy range, so that the light curve is representative of the flux of the target in that energy range. This is also important for selecting energy bands for computing hardness ratios. The 'chmin' and 'chmax' parameters are hidden parameters of the task 'pcaextlc2' which allows you to choose the channel range of interest. Since these are hidden parameters, you need to specify them explicitly on the command line when you call 'pcaextlc2': pcaextlc2 ... chmin=5 chmax=30 Note that 'chmin' and 'chmax' refer to the channel assignments in the original 0-249 channel range, not the 0-128 range of Standard2. See the energy to channel table in order to estimate which channel values to use. Hardness ratios are ratios of count rates in different energy bands. Different researchers have different meanings for the term "hardness ratio," so it is important to be clear which ratio is intended. In either case, one must extract more than one light curve from the same input data using exactly the same time filtering settings. It is important to have the same time filtering settings (GTI file, timemin, timemax, etc) so that the light curve sample values in different energy ranges match up row-by-row. For example, these settings could be used: pcaextlc2 ... outfile=93067-soft.lc chmin=5 chmax=30 pcaextlc2 ... outfile=93067-hard.lc chmin=31 chmax=60 and now 93067-soft.lc will be in the soft band (channels 5-30, approximately 2-10 keV) and 93067-hard.lc will be in the hard band (channels 31-60, approximately 10-20 keV). Hardness ratios can be computed by hand. The task 'ftpaste' can be used to merge two light curve files in different energy bands, and then ftcalc can be used to compute the ratio. The task 'lcurve' is also commonly used to combine light curves in order to calculate hardness ratios. In order to use this approach, one needs to enter a @filelist.lis input file, which contains two input series. Recommendations for PCU and Layer SelectionsThe subsection has a few recommendations about which PCU detectors and/or layers to choose using the 'pculist' and 'layerlist' parameters. This selection can make a difference in some cases. Each PCU has a slightly different mechanical alignment with respect to the pointing axis of the observatory, and thus samples a slightly different position in a PCU's collimator response. The light curves produced by 'pcaextlc2' are not collimator corrected. What this means is that the light curve count rate versus flux conversion scale is slightly different for each PCU (<8%). Thus it is usually recommended to only use a single PCU when extracting light curves. Since PCU2 was enabled for every observation, and never had propane or breakdown problems, it is usually advisable to set pculist=2 for light curve extractions. For faint sources, fainter than about 10 mCrab = 2 x 10-10 erg s-1 cm-2 = 20 ct/s/PCU, the differences due to collimator response differences are smaller than the Poisson counting error, and so there may be some advantage to including more PCUs to reduce the Poisson counting error. This is a scientific judgement call. Also for faint sources, most of the photons will be absorbed in the top layer, and not many source counts will survive layer 2 or layer 3. Layers 2 and 3 will likely be background dominated and will contribute more noise than signal to your results. Therefore, the PCA team typically recommends that for targets <10-20 mCrab, use only layer 1 ("layerlist=1"). By default, data from PCUs 0 and 1 are excluded by the software discussed here, after those detectors lost their propane layer. The PCA team does not normally recommend analyzing this data without great care. Details of Light Curve ExtractionThe input to this tool is a set of PCA Standard2 files. To gain full benefit of this tool, the user must provide dead-time calculated version of the Standard2 file. For beginning users, the output files of the 'pcaprepfile2' task are sufficient. For advanced users, the equivalent output of 'pcadeadcalc2' is acceptable. If background estimates are required, the user must background files, also with dead-time quantities calculated. The output of the task is a light curve file (.lc file) which contains the source count rate in the RATE column Using the default settings to this task, the output light curve file has rates that are dead-time corrected, and contain only counts and exposure from live detectors, as described in the help file for the task 'pcadeadcalc2'. If lcmode=RATE, the output is a RATE column (counts per live detector per unit live time), and the ERROR column represents the Poisson rate error. If lcmode=COUNTS, the output is a COUNTS column (total counts during each time bin), ERROR is the Poisson counting error. For lcmode=COUNTS, the FRACEXP value can be greater than 1.0, indicating more than one PCU is enabled. In this case, the rate per live PCU can be computed as COUNTS/TIMEDEL/FRACEXP. For dead-time purposes, if deadcorrtype=LIVE then the live-time-corrected values of the light curve samples is computed, and the live time is reported in the column LiveTime. Dead-time corrected samples are indicated by the presence of the DEADAPP='T' keyword. If deadcorrtype=ON, then the light curve samples are corrected for number of PCUs enabled (i.e. "on-time"), but not for dead-time. The on-time is then reported in the OnTime column. If only correcting for on-time, then DEADAPP='F'alse. For background subtraction purposes, a set of background estimates must be supplied in the bkg_infile parameter. If bkgsub=NO, then background values are produced and reported in the BACKV and BACKE columns, but they are not subtracted. If bkgsub=YES, then the background is subtracted, i.e. RATE is recomputed as (RATE-BACKV), and ERROR is recomputed as SQRT(ERROR*ERROR+BACKE*BACKE). The presence of background subtraction is indicated by the BACKAPP='T' keyword. Advanced UsersAdvanced users can still use the more basic tools for fine-grained control instead of this task. Because extraction and dead-time correction require the user to perform several careful operations, the use of this tool is recommended to make sure that the operations are done consistently. Generally, the PCA team and the RXTE GOF recomend to not change the default for the advance parameters of this tool without specific need. Advanced users that wish to tweak their settings are not out of luck. The archane settings to 'xtefilt' and 'pcabackest' are still available as hidden parameters. By typing 'fhelp pcaprepobsid' one can find all of these hidden settings. However, for 99% of use cases, this should not be necessary. The Next StepsAt this point, you should have standard light curve and spectral data products from your RXTE obsevation. What do you do with them? For light curves, you may wish to simply lot the light curve, for which you can use the 'fv' package included with HEASoft. For more technical analysis of light curves, such as hardness ratios or Fourier power spectra, the 'Xronos' package included with HEASoft may be appropriate. For spectra, we recommend to use the 'XSPEC' package included with HEASoft, but any spectral analysis package such as 'ISIS' will be sufficient. Last modified: Fri Aug 23 10:41:33 EDT 2019 |