THE XMM-NEWTON ABC GUIDE, STREAMLINED
EPIC-MOS (TIMING mode), SAS GUI
Contents
Prepare the Data
Reprocess the Data
Introduction to xmmselect
Apply Standard Filters
Make a Light Curve
Extract the Source and Background Spectra
Check for Pile Up
Determine the Spectrum Extraction Areas
Create the Photon Redistribution Matrix (RMF) and Ancillary File (ARF)
Prepare the Data
Please note that the two tasks in this section (cifbuild and odfingest) must be run in the ODF directory. These are the only tasks with that requirement, and after this section, we will work exclusively in our reprocessing directory.Many SAS tasks require calibration information from the Calibration Access Layer (CAL). Relevant files are accessed from the set of Current Calibration File (CCF) data using a CCF Index File (CIF).
To make the ccf.cif file, first make sure the environment variables are set:
cd ODF setenv SAS_ODF /full/path/to/ODF/directory/ setenv SAS_ODFPATH /full/path/to/ODF/directory/
Next, call cifbuild from the SAS GUI. A window with the parameter options will appear; the defaults should be fine, so just click "Run".
To use the updated CIF file in further processing, you will need to reset the environment variable SAS_CCF:
setenv SAS_CCF /full/path/to/ODF/ccf.cifThe task odfingest extends the Observation Data File (ODF) summary file with data extracted from the instrument housekeeping data files and the calibration database. It is only necessary to run it once on any dataset, and will cause problems if it is run a second time. If for some reason odfingest must be rerun, you must first delete the earlier file it produced. This file largely follows the standard XMM naming convention, but has SUM.SAS appended to it. After running odfingest, you will need to reset the environment variable SAS_ODF to its output file. To run odfingest and reset environment variable, call odfingest from the SAS GUI. As before, a pop-up window with the parameter options will appear; the defaults should be fine, so just click "Run". (It is safe to ignore the warnings.)
To change the environmental variable, type
setenv SAS_ODF /full/path/to/ODF/full_name_of_*SUM.SASYou will likely find it useful to alias these environment variable resets in your login shell (.cshrc, .bashrc, etc.).
Reprocess the Data
To reprocess the data, make a new working directory and call emproc from it:cd .. mkdir PROC cd PROCThen, close and re-open the SAS GUI, so that it will place the output files in the new directory, and call emproc. The defaults are fine for most observations, so just click "Run". (It is safe to ignore the warnings.)
By default, the task does not keep any intermediate files it generates. Emproc designates its timing output event files with "TimingEvts.ds". It is convenient to rename the event file something easy to type:
cp 0201_0070740101_EMOS1_S001_TimingEvts.ds mos1_te.fits
If you are likely to want to extract a background spectrum for your source, you will also need to consider the imaging event file. We might as well deal with that while we're here. Remember that whatever filtering is done on the timing event file must also be done on the image event file.
cp 0201_0070740101_EMOS1_S001_ImagingEvts.ds mos1_ie.fits
Introduction to xmmselect
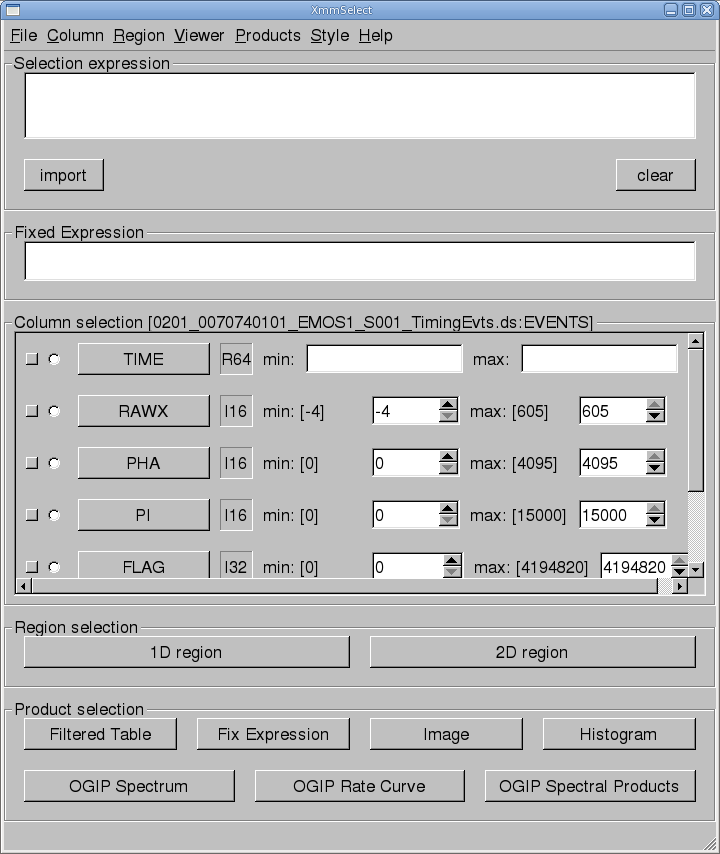
The task xmmselect is used for many procedures in the GUI. Like all tasks, it can easily be invoked by starting to type the name and pressing enter when it is highlighted.When xmmselect is invoked a dialog box will first appear requesting a file name. You can either use the browser button or just type the file name in the entry area, "mos1_te.fits:EVENTS" in this case. To use the browser, select the file folder icon; this will bring up a second window for the file selection. Choose the desired event file, then the "EVENTS" extension in the right-hand column, and click "OK". The directory window will then disappear and you can click "Run" on the selection window.
When the file name has been submitted the xmmselect GUI will appear, along with a dialog box offering to display the selection expression; this is shown in Figure 1. The selection expression will include the filtering done to this point on the event file, which for the pipeline processing includes for the most part CCD and GTI selections.
Different event files can be loaded into xmmselect by going to "File -> New Table".
Apply Standard Filters
The filtering expression for the MOS in TIMING mode is:
(PATTERN <= 12)&&(PI in [200:12000])&&#XMMEA_EM
The first two expressions will select good events with PATTERN in the 0 to 12 range. The PATTERN value is similar the GRADE selection for ASCA data, and is related to the number and pattern of the CCD pixels triggered for a given event. Single pixel events have PATTERN == 0, while double pixel events have PATTERN in [1:4] and triple and quadruple events have PATTERN in [5:12].
The second keyword in the expressions, PI, selects the preferred pulse height of the event. For the MOS, it should be between 200 and 12000 eV. This should clean up the image significantly with most of the rest of the obvious contamination due to low pulse height events. Setting the lower PI channel limit somewhat higher (e.g., to 300 or 400 eV) will eliminate much of the rest.
Finally, the #XMMEA_EM filter provides a canned screening set of FLAG values for the event. (The FLAG value provides a bit encoding of various event conditions, e.g., near hot pixels or outside of the field of view. Setting FLAG == 0 in the selection expression provides the most conservative screening criteria and usually is not necessary for the MOS.)
To filter the data using xmmselect,
- Enter the filtering criteria in the "Selection Expression" area at the top of the xmmselect window:
(PATTERN <= 12)&&(PI in [200:12000])&&#XMMEA_EM
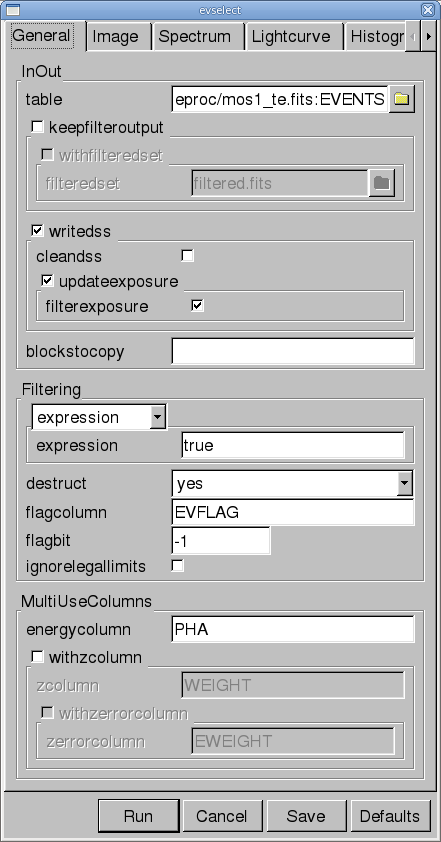
- Click on the "Filtered Table" box at the lower left of the xmmselect GUI. This will bring up the evselect GUI.
- In the "General" tab, change the filteredset parameter, the output file name, to something useful, e.g., mos1_te_filt.fits.
- Click "Run".
Remember to do the same for the image event file if you plan to use it.
Make a Light Curve
Sometimes, it is necessary to use filters on time in addition to those mentioned above. This is because of soft proton background flaring, which can have count rates of 100 counts/sec or higher across the entire bandpass. To determine if our observation is affected by background flaring, we can make a light curve by calling xmmselect and loading the event file as shown above. Then,
- Check the round box to the left of the "Time" entry.
- Click on the "OGIP Rate Curve" button near the bottom of the page. This brings up the evselect GUI (see Figure 2).
- Click on the "Lightcurve" tab and change the "timebinsize" to a reasonable amount, e.g. 20 s. In the "rateset" textbox, enter the name of the output file, for example, mos1_ltcrv.fits.
- Click on the "Run" button at the lower left corner of the evselect GUI.
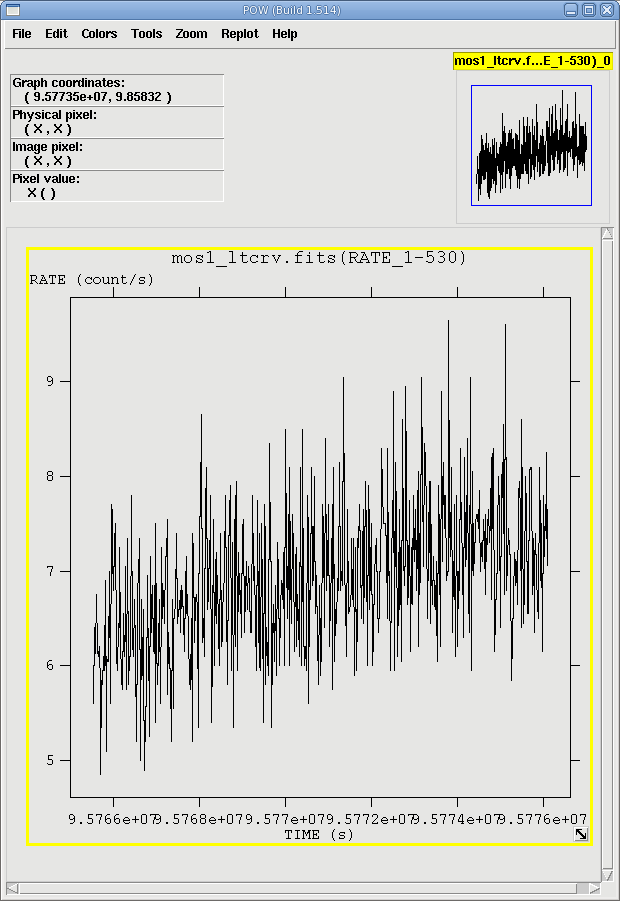
The output file mos1_ltcrv.fits is automatically displayed in Grace. It can also be viewed by using fv and is shown in Figure 3. No flares are evident, so we will continue to the next section. However, if a dataset does contain flaring, it should be removed in the same way as shown for EPIC IMAGING mode data here.
- fv mos1_ltcrv.fits &
Extract the Source and Background Spectra
First, we will need to make an image of the filtered event file. We will load the filtered event file into xmmselect, and then,
- Make an image by checking the square boxes to the left of the "RAWX" and "TIME" entries to indicate which is on the X and Y axis. Click the "Image" button near the bottom of the page. This brings up the evselect GUI.
- In the "Image" tab, enter the name of the output file in the imageset box; we will use mos1_te_image.fits. Toggle Binning to binSize, and confirm that ximagebinsize and yimagebinsize are set to 1.
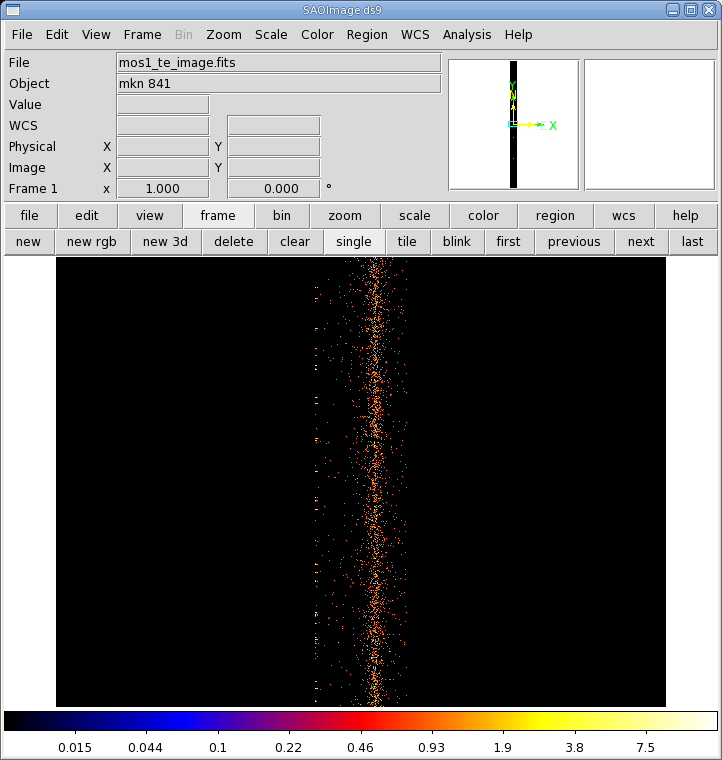
- Click the "Run" button on the lower left corner of the evselect GUI. The image will be displayed automatically in a ds9 window.
The image is shown in Figure 4. The source is centered on RAWX=314. We will extract this and the 10 pixels on either side of it, keeping only the highest quality, single pattern events (FLAG==0 && PATTERN==0)).
- Enter the filtering criteria in the "Selection Expression" area at the top of the xmmselect window: (FLAG==0) && (PATTERN==0) && (RAWX in [304:324]).
- Click the round button next the PI column on the xmmselect GUI
- Click on "OGIP Spectrum".
- In the "General" tab, check keepfilteroutput and withfilteredset. In the filteredset box, enter the name of the event file output. We will use mos1_filt_source.fits.
- In the "Spectrum" tab, set the file name and binning parameters for the spectrum. Confirm that withspectrumset is checked. Set spectrumset to the desired output name. We will use source_pi.fits. Confirm that withspecranges is checked. Set specchannelmin to 0 and specchannelmax to 11999.
- Click "Run".
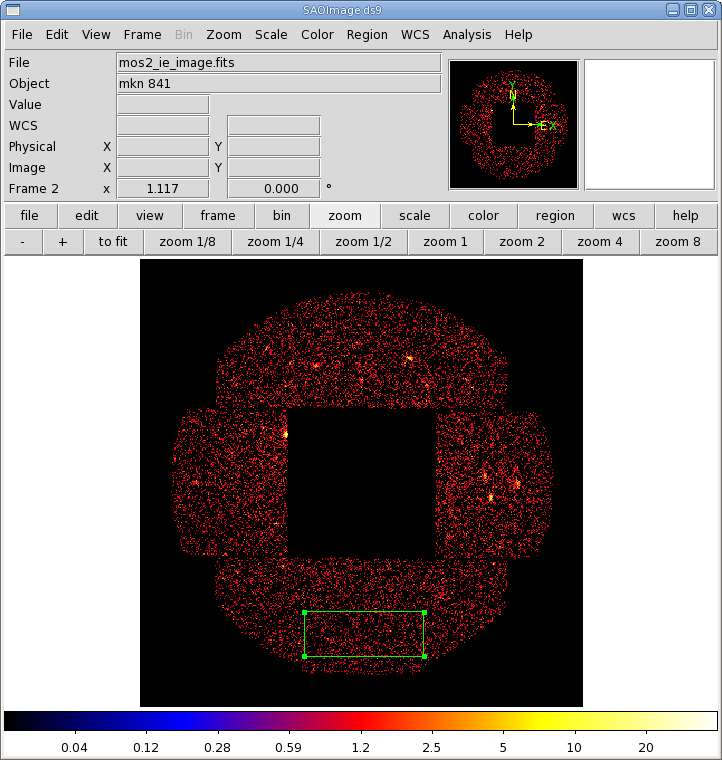
If needed, we can also extract a background spectrum. For this, we will use the imaging event list, since we want the background to be as far away from the source as possible. As with the source spectrum, we will need to make an image first. We will load the filtered image event file mos1_ie_filt.fits into xmmselect, and then,
- Make an image by checking the square boxes to the left of the "DETX" and "DETY" entries to indicate which is on the X and Y axis. Click the "Image" button near the bottom of the page. This brings up the evselect GUI.
- In the "Image" tab, enter the name of the output file in the imageset box; we will use mos1_ie_image.fits. Toggle Binning to binSize, and let ximagebinsize and yimagebinsize be set to 100.
- Click the "Run" button on the lower left corner of the evselect GUI. The image will be displayed automatically in a ds9 window.
The image is shown in Figure 5, with the background extraction region overlayed. Once again, we will only keep the high quality events, but we'll loosen the restriction on the event pattern a bit, using patterns 0, 1, and 3, since we're using the outer CCDs.
- Enter the filtering criteria in the "Selection Expression" area at the top of the xmmselect window: (FLAG==0) && (PATTERN==0) && ((DETX,DETY) in BOX(301.5,-13735.5,10700,4000,0))
- Click the round button next the PI column on the xmmselect GUI
- Click on "OGIP Spectrum".
- In the "General" tab, check keepfilteroutput and withfilteredset. In the filteredset box, enter the name of the event file output. We will use mos1_filt_bkg.fits.
- In the "Spectrum" tab, set the file name and binning parameters for the spectrum. Confirm that withspectrumset is checked. Set spectrumset to the desired output name. We will use bkg_pi.fits. Confirm that withspecranges is checked. Set specchannelmin to 0 and specchannelmax to 11999.
- Click "Run".
|