Hitomi Archive: Getting started
General
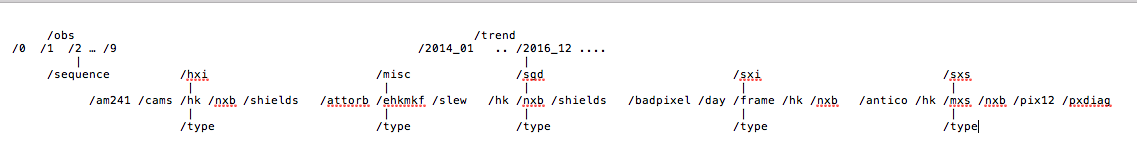
The Hitomi archive is divided in two main directories science and trend data. The science
data are organized by sequence, where a sequence corresponds to an observation of one day
in lenght. A sequence contains data from all instruments and is identified by a 9 digit number.
The /obs directory contains the science data and it is subdivided in directories named
after the first digits of the sequence number under which the observations are located.
The trend area instead is divided in months interval named YYYY_NN, where YYYY is the year
and NN is the numeric value for the month. For each month the trend (or monitor) data are organized
by type for each of the instrument. The archive structure is therefore:
Archive access : Web, wget or download script
At the HEASARC the Hitomi archive can be accessed using the web interfaces
Browse , wget or with a download script.
- Access via Web : The archive access via the Web makes use of database table to search for the science data.
The database table is hitomaster
which records high-level information for each Hitomi observation.
This table can be queried for position, time and several other parameters, and from the result page
it is possible to select and retrieve the data files.
- Wget : If you have wget
installed, and the observation id is already known, the data can be retrieved via 'wget'.
For example using the HHTP protocal, if the user wants to retrieve the sequence '100001010' observed
on Feb 24 2016, she/he can retrive the entire sequence (see Example 1) or data for a single instrument
(see Example 2)
Example 1
wget -q -nH --cut-dirs=5 -r -l0 -c -N -np -R 'index*' -erobots=off --retr-symlinks https://heasarc.gsfc.nasa.gov/FTP/hitomi/data/obs/1/100040010/
Example 2
wget -q -nH --cut-dirs=5 -r -l0 -c -N -np -R 'index*' -erobots=off --retr-symlinks https://heasarc.gsfc.nasa.gov/FTP/hitomi/data/obs/1/10004010/sxi/
-
download_wget : donwload_wget is a PERL script that
uses the wget protocol, similarly to the example above, but simplify the command and allow for more flexility.
You can retrive the
download_wget and place it in your local machine
Example: To download a specific sequence type:
> download_wget.pl https://heasarc.gsfc.nasa.gov/FTP/hitomi/data/obs/1/100001010
To get help type : > download_wget.pl help
|



