Next: 5. XRISM Data Analysis Up: xrism abc Previous: 3. Analyses Tools and Contents
This chapter describes the XRISM observation data, including the directory structure, data files, and their formats. The data format and structure are similar to some X-ray observatory mission data (e.g., ASCA, Suzaku) with minor variations.
Unique 9-digit sequence numbers are used to organize the XRISM observations. A sequence number is assigned for all data during an observation and during the slew from the previous observation. The initial data processing at JAXA and NASA, hereafter pipeline, collects all relevant data of an observation, calibrates and screens the data, makes standard analysis for quick looks, and stores all products in a package with a sequence number. NASA and JAXA have their own data archives open to the public through the following URLs.
NASA: https://heasarc.gsfc.nasa.gov/FTP/xrism/data/obs/
JAXA: https://data.darts.isas.jaxa.jp/pub/xrism/data/obs/
Users can directly access these URLs to download any packages, but they can find the desired datasets more easily through the archival portal sites (see Section 5.2).
A XRISM data package is stored under a directory with its sequence number name. There are four directories under the top directory.
Each of the Resolve and Xtend directories has the following four sub-directories.
All science and HK files are in FITS format, while processing logs are in plain text.
Users may use the following files in downstream software commands.
| attitude | auxil/xaOBSID .att |
| filter | auxil/xaOBSID .ehk |
| orbit | auxil/xaOBSID .orb |
| unfiltered event | (resolve or xtend)/event_uf/xaOBSID iii_P0mmmmmm_uf.evt |
| cleaned event | (resolve or xtend)/event_cl/xaOBSID iii_P0mmmmmm_cl.evt |
where
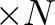
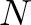 =6 for Resolve, =8 for Xtend)
=6 for Resolve, =8 for Xtend)
The instrument mode describes the applied X-ray filter in use for Resolve. The former 3 digits of the instrument mode describe the applied CCD window and burst mode in use for Xtend.
| Resolve | |
| Sub-string | X-ray Filter |
| px0000 | Undefined |
| px1000 | OPEN |
| px2000 | Al/Polyimide |
| px3000 | Neutral Density (ND) |
| px4000 | Be |
| px5000 | Fe 55 calibration source |
| Xtend | |
| Sub-string | CCD Window/Burst Mode |
| 300 | all four CCDs in full window mode |
| 311 | CCD1 & CCD2 in 1/8 window mode |
| 312 | CCD1 & CCD2 in full window + 0.1 sec burst mode |
| 313 | CCD1 & CCD2 in 1/8 window + 0.1 sec burst mode |
| 320 | CCD3 & CCD4 in full window mode |
All files are compressed in gzip, so they have extra .gz extensions. FTOOLS software understands gzipped files, so users do not need to unzip them.
The pipeline produces two types of event files for each instrument: the 'unfiltered' file (*_uf.evt) under the event_uf directory and the 'cleaned' file (*_cl.evt) under the event_cl directory. The unfiltered files contain all events telemetered from the observatory; the events are calibrated but not screened. The pipeline at the data center or the xapipeline reprocessing tool feeds these unfiltered files, filters events that satisfy specific conditions (e.g., event type, grades) or time intervals (e.g., outside of South Atlantic Anomaly) and outputs 'cleaned' event files. The screening applied by the pipeline is the minimum set of filters considered to be compatible with all observations, including those of bright sources. The archival cleaned files and those reprocessed with xapipeline with the default setup use the screening criteria defined in Table 5.1 for Resolve data and Table 5.2 for Xtend data, which shows the minimum screening. Users can adjust the screening conditions to optimize for their scientific goals following the guidance of Section 6.3 for Resolve and Section 7.3 for Xtend at their own risk. Users should always use cleaned event files for science analysis.
The pipeline uses various coordinate systems (RAW, ACT, DET, FOC, SKY) to map detected event locations on the detectors to celestial coordinates. Event files store all coordinate values for each event, but general users need only the following coordinates for science analysis. See also Fig. 5.1 for Resolve and Fig. 6.2 for Xtend in POG.
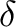 ) increases
in the +Y direction and Right Ascension (
) increases
in the +Y direction and Right Ascension (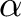 ) increases in the -X direction —
incremented every
) increases in the -X direction —
incremented every 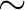 1.77 arcsec.
Their origin is at the celestial reference coordinates in the HEADER keywords (TCRVL31/TCRVL32),
shared between the Resolve and Xtend data of the same observation.
A detector pixel can generally be mapped onto multiple SKY pixels, so the software picks a SKY pixel for each event based on its weighted positional probability and stores the values in the X/Y columns of the event FITS files.
Standard image viewers such as ds9 and Ximage can automatically
convert the SKY coordinates to the world coordinate system.
1.77 arcsec.
Their origin is at the celestial reference coordinates in the HEADER keywords (TCRVL31/TCRVL32),
shared between the Resolve and Xtend data of the same observation.
A detector pixel can generally be mapped onto multiple SKY pixels, so the software picks a SKY pixel for each event based on its weighted positional probability and stores the values in the X/Y columns of the event FITS files.
Standard image viewers such as ds9 and Ximage can automatically
convert the SKY coordinates to the world coordinate system.
 6 pixel array in the positive quadrant,
with the missing corner pixel for calibration at (1, 1), and increments the coordinates by 1
with every pixel. The Xtend DET coordinate system originates at the corner of
the CCD array on CCD_ID=1, having the x- and y-axes of the vertical and horizontal CCD pixel directions of CCD_ID=1. The four CCDs do not perfectly align, so the DETX/Y axes are not
strictly parallel to the CCD rows or columns of the other CCDs (CCD_IDs = 0, 2, 3).
The event FITS files store these values in the DETX/DETY columns.
6 pixel array in the positive quadrant,
with the missing corner pixel for calibration at (1, 1), and increments the coordinates by 1
with every pixel. The Xtend DET coordinate system originates at the corner of
the CCD array on CCD_ID=1, having the x- and y-axes of the vertical and horizontal CCD pixel directions of CCD_ID=1. The four CCDs do not perfectly align, so the DETX/Y axes are not
strictly parallel to the CCD rows or columns of the other CCDs (CCD_IDs = 0, 2, 3).
The event FITS files store these values in the DETX/DETY columns.
Each event has a PI (pulse invariant) value that is proportional to the incident photon energy. This value is calculated from the sensor's pulse outputs (pulse height value: PHA for Resolve, PHAS for Xtend) and adjusted to the linear energy scale of the incident photon using the calibration information. Xselect produces a PI value histogram (event counts per each PI value or channel) from the selected events for spectral analysis. Xspec fits the histogram with physical models with the corresponding detector response.
Each event has a TIME value, which describes the event detection time in seconds from the XRISM mission time origin (2019-01-01 00:00:00 UTC). The mission time origin is stored in the MJDREFI and MJDREFF keywords of all XRISM FITS file headers, so HEASoft or OGIP-compliant timing software can convert the mission time to a human-friendly time system. Users can also convert XRISM time to various time systems on the xTime HEASARC web page.
By default, XSELECT (see Section 3.1.3) uses the following setups for XRISM data analyses4.1. Users can change them during XSELECT sessions after loading event data.
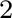 .
.
 1 value.
1 value.
| Column | Description | Range |
| ITYPE/TYPE | Resolution Grade | 0 7/Xx 7/Xx |
| 0/Hp: High-resolution Primary | ||
| 1/Mp: Mid-resolution Primary | ||
| 2/Ms: Mid-resolution Secondary | ||
| 3/Lp: Low-resolution Primary | ||
| 4/Ls: Low-resolution Secondary | ||
| 5/Bl: Baseline event (diagnostic) | ||
6/El: Lost event
 |
||
| 7/Rj: rejected events | ||
| PIXEL | Pixel number | 0 35 35 |
| RISE_TIME | Event pulse time from the baseline to the peak | 0 255 255 |
| PI | Pulse Invariant energy channel | 0 59999 59999 |
STATUS[ ] ] |
16 bit Event Flag (14 in use) | b0: no |
| [1]: outside of all-pixel GTIs | b1: yes | |
| [2]: outside of individual-pixel GTIs | ||
| [3]: coincidence with anti-co event | ||
| [4]: coincidence with event on any pixel except 12 | ||
| [5]: coincidence with pixel 12 event | ||
| [6]: [5] & passed energy test for absorption of electron ejected | ||
| from 12 | ||
| [7]: candidate electrical crosstalk event or its source | ||
| [8]: [7] & largest PHA of coincident group | ||
| [9]: during pulse of MXS direct source | ||
| [10]: during afterglow of MXS direct source | ||
| [11]: during pulse of MXS indirect source | ||
| [12]: during afterglow of MXS indirect source | ||
| [13]: event likely contaminated by untriggered electrical crosstalk | ||
| [14]: [13] & largest PHA of coincident group | ||
| ITYPE/TYPE provides a numerical/adjectival representation of the event grade. It is also | ||
| conventionally called GRADE. | ||
 The events the onboard software is unable to process within the available computational The events the onboard software is unable to process within the available computational |
||
| time. | ||
| Column | Description | Range |
| GRADE | 5 5 pixel pulse height patterns 5 pixel pulse height patterns |
0 11 11 |
0: Single
 |
||
| 1: Corner split | ||
2: Vertical split
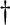 |
||
3: Left split
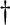 |
||
4: Right split
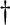 |
||
| 5: Vertical, left, or right split with a corner | ||
6: 2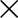 2 box split with a low corner pulse-height 2 box split with a low corner pulse-height
 |
||
7: 3 pixels vertical or horizontal split with an outer 5 5 pixel 5 pixel |
||
| above thres. | ||
| 8: unused | ||
| 9: 3 pixels vertical or horizontal split with no outer 5x5 pixel | ||
| above thres. | ||
| 10: GRADE 2, 4, 5, or 6 with an outer 5x5 pixel above thres. | ||
11: 2 2 box split with a high corner pulse-height 2 box split with a high corner pulse-height |
||
| PI | Pulse Invariant energy channel | 0 4095 4095 |
STATUS[ ] ] |
48 bit Event Flag (37 in use) | b0: no |
| [1]: Bad event based on all other status flags | b1: yes | |
| [2]: Inside the calibration source region | ||
[3]: Outside of CCD |
||
[4]: Outside of window |
||
[5]: Outside of the area discrimination |
||
[6]: Charge Injection (CI) row |
||
[7]: Bad pixel from CALDB |
||
[8]: Bad column from CALDB |
||
[9]: Hot pixel from pre-pipeline |
||
[10]: Flickering pixel detected by searchflickpix |
||
[11]: CCD boundary |
||
[12]: Window boundary |
||
| [13]: Segment boundary | ||
| [14]: Area discrimination boundary | ||
| [15]: At least one 3x3 surrounding pixel has bad status | ||
[16]: CI trailing row |
||
[17]: CI preceding row |
||
[18]: Preceding/following of bad column |
||
[19]: Neighbor of bad/hot pixel and bad column |
||
[20]: Neighbor of flickering pixel |
| Column | Description | Range |
STATUS[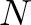 ] ] |
[21]: Neighbor of preceding/following of bad column STATUS | b0: no |
| [22]: Neighbor of CCD/window boundary | b1: yes | |
| [23]: Neighbor of segment boundary | ||
| [24]: 3x3 info is present but 5x5 is absent | ||
[25]: 3x3 is absent |
||
[26]: PHAS[0] 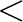 event threshold event threshold |
||
[27]: Video temperature is out of range |
||
[28]: Lack of video temp HK at time close to the event |
||
| [29]: Correction value is negative | ||
[30]: Null value by correction process |
||
| [31]: 1st trailing row of the CI rows | ||
| [32]: 1st preceding row of the CI rows | ||
| [33]: 2nd trailing row of the CI rows | ||
| [34]: 2nd preceding row of the CI rows | ||
| [35]: 3rd trailing row of the CI rows | ||
| [36]: 3rd preceding row of the CI rows | ||
| [37]: Cosmic ray echo pixel | ||
 Grades corresponding to X-ray (as opposed to cosmic ray) events. Grades corresponding to X-ray (as opposed to cosmic ray) events. |
||
 Bad pixel events removed with the standard screening. Bad pixel events removed with the standard screening. |
||