Next: 6. Resolve Data Analysis Up: xrism abc Previous: 4. XRISM Data Specifics Contents
This chapter briefly outlines the initial analysis steps common to Resolve and Xtend. Subsequent chapters 6 & 7 describe the following steps specific to each instrument.
This and the following chapters use the XRISM first light observation data of the supernova remnant N132D
(Obs_ID = 000126000) as command examples.
The object is moderately extended by 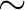 1 arcmin, but we sometimes treat the source
as a point source.
Users can freely change the command options with “N132D” to other arbitrary names
while replacing those with 000126000 to the corresponding files in their data packages.
1 arcmin, but we sometimes treat the source
as a point source.
Users can freely change the command options with “N132D” to other arbitrary names
while replacing those with 000126000 to the corresponding files in their data packages.
Users need HEASoft version 6.34 or a later version, XRISM CALDB, and the SAOImage ds9 viewer. Read the Software chapter in Section 3 for the details.
Most XRISM Guest Observer (GO) data are proprietary for 1 year after being fully processed. During the proprietary period, the data are downloadable but encrypted with gpg encryption. Only the observation's contact person receives the instructions for and decrypting the data from the Science Data Center, or SDC. After the proprietary period, all data are decrypted and available to anybody.
Non-GO users can search and access public data through the two web portals at NASA's HEASARC and ISAS/JAXA's DARTS.
https://heasarc.gsfc.nasa.gov/docs/archive.html
They provide multiple options for data downloads. Below is an example of downloads from NASA and JAXA through Unix terminal commands. Similar commands are in the instructions for GO observers.
NASA:
term> wget -nv -m -np -nH –cut-dirs=5 -R "index.html*" –execute robots=off
–wait=1 https://heasarc.gsfc.nasa.gov/FTP/xrism/data/obs/0/000126000 /
JAXA:
term> wget -nv -m -np -nH –cut-dirs=6 -R "index.html*" –execute robots=off
–wait=1 https://data.darts.isas.jaxa.jp/pub/xrism/data/obs/rev3/0/000126000 /
The last directory but one, with “0” in the above examples, organizes the observation type
with a single-digit number (0 9) (2 for GO observations).
Users should not omit “/” at the end of the html address,
or they copy many empty directories at the same directory level.
9) (2 for GO observations).
Users should not omit “/” at the end of the html address,
or they copy many empty directories at the same directory level.
The XRISM operations team makes logs of satellite and instrument anomalies, which are available from the following web page.
https://xrism.isas.jaxa.jp/research/observers/operation_log/index.html
Unusual data are often associated with these events.
The pipeline processing produces the Energy Scale Quality Reports for each observation, which summarizes the Resolve per-pixel and full-array energy resolution and gain accuracy based on Filter Wheel and pixel 12 Fe55 Calibration source data analysis. Users can download the reports from the following site.
https://heasarc.gsfc.nasa.gov/docs/xrism/analysis/gainreports/index.html
Users are advised to check these notes before analyzing the data.
The XRISM pipeline generates quick-look preview products under each instrument's products directory (see Section 4.2). This directory includes images in multiple energy bands and the main target's light curves and spectra (whole sensor for Resolve, within 2.5 arcmin from the on-axis position for Xtend) in FITS or GIF format. These products are handy for checking the quality of the data obtained. However, we strongly discourage using them for detailed science or calibration analysis. The directory does not contain spectral arf responses for either instrument, so users cannot perform spectral fits with these products.
The instrument teams regularly monitor the instrumental responses and their time evolutions and update the results in CALDB, sometimes promptly, if their findings significantly impact the scientific outcome. Data downloded a while ago may well be outdated. Also, the Science Data Center (SDC) does not reprocess archival data for each CALDB release, so archival data placed a while ago may not also use the latest calibration. Users are encouraged to check the calibration information and apply the latest calibration to the analysis dataset.
Users can find the latest XRISM calibration information on the following calibration web page.
https://heasarc.gsfc.nasa.gov/docs/xrism/calib/index.html
All FITS files have the following keywords, which record the calibration version applied by the pipeline.
| PROCVER: | processing script version |
| SOFTVER: | software package version used for the pipeline |
| CALDBVER: | calibration version used for the pipeline |
If the calibration is outdated, users should apply the latest calibration to the data with the HEASoft XRISM task xapipeline. This script runs the following three stages:
Here is a command example.
term> xapipeline indir=000126000 outdir=000126000 _rep
steminputs=xa000126000
stemoutputs=DEFAULT entry_stage=1
exit_stage=2 instrument=ALL verify_input=no
This command runs in the directory with the downloaded data's top directory (000126000 ) and outputs all reprocessed data to the directory, 000126000 _rep5.1. Users can control the reprocessing stages with the entry_stage and exit_stage options. For example, a run with entry_stage=2 does not re-calibrate data, while a run with exit_stage=3 also makes quick-look products. Users can reprocess only one instrument data with the instrument option. xapiepline has many command options to control all the tasks running under it. To get a list of all the parameters, please check the command line manual by typing
term> fhelp xapipeline
or visit the online help.
Tables 5.1 and 5.2 show the Resolve and Xtend standard screening criteria the pipeline uses to produce cleaned event files. There are two types of data screening: one with time and one with event characteristics. The time screening excludes intervals not suitable for the target's science study — the slew from the previous target, the Earth occultation of the target, the South Atlantic Anomaly (SAA) passages with high particle background, the Resolve ADR cooling cycles with significant spectral gain uncertainty, and event losses during the onboard data processing and telemetry. The files in the reference column record these time intervals, and the pipeline excludes them to include only “good time interval” or GTI. The data obtained during the GTI intervals still include particle background or spurious events. The event screening selects events that maximize X-ray signals for most objects.
| Screen | Reference | Condition | Comments |
| Time | .ehk | ANG_DIST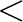 1.5 1.5 |
pointing target |
| SAA_SXS==0 | outside SAA | ||
ELV 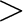 5 5 |
above the Earth rim | ||
DYE_ELV 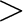 5 5 |
above the Bright Earth rim | ||
| Time | _gen.gti | inside GTIPOINT | excl. slew |
| inside GTIATT | excl. bad attitude | ||
| Time | .mkf | nominal instrument status | |
| Time | rsl_tel.gti | inside GTIADROFF | excl. ADR cycle |
| Time | _el.gti | inside GTILOSTOFF | excl. lost event interval
 |
| Event | _uf.evt | ITYPE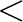 5 5 |
|
(SLOPE_DIFFER==b0 PI PI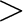 22000) 22000) |
|||
| QUICK_DOUBLE==b0 | |||
| STATUS[2]==b0 | |||
| STATUS[3]==b0 | excl. antico events | ||
| STATUS[6]==b0 | excl. e- from pix 12 | ||
RISE_TIME 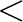 127 127 |
|||
| PIXEL!=12 | excl. calibration pixel | ||
 8 8 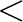 TICK_SHIFT TICK_SHIFT 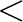 7 7 |
|||
| The screening formulae connect these conditions with AND unless explicitly stated. | |||
| Strings in the Reference column come after “xaOBSID.” | |||
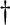 applied to antico events only. applied to antico events only. |
|||
| Screen | Reference | Condition | Comments |
| Time | .ehk | ANG_DIST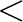 1.5 1.5 |
pointing target |
| SAA_SXI==0 | outside SAA | ||
T_SAA_SXS 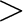 277 277 |
Time after SAA passage | ||
ELV 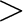 5 5 |
above the Earth rim | ||
DYE_ELV 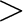 20 20 |
above the Bright Earth rim | ||
| Time | _gen.gti | inside GTIPOINT | excl. slew |
| inside GTIATT | excl. bad attitude | ||
| Time | .mkf | SXI_USR_CCD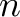 _OBS_MODE==1 _OBS_MODE==1 |
nominal instrument status |
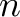 : appropriate CCD_IDs : appropriate CCD_IDs |
|||
| Time | sxi_tel.gti | inside GTITEL | excl. times of telemetry saturation |
| Time | _uf.evt | inside GTIEVENT | inc. GTIs of the event data |
| Event | _uf.evt | GRADE==0,2,3,4,6 | |
| STATUS[1]==b0 | |||
PROC_STATUS[1,2]==b0
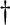 |
|||
| The screening formulae connect these conditions with AND unless explicitly stated. | |||
| Strings in the Reference column come after “xaOBSID.” | |||
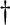 PROC_STATUS reports errors found in the telemetry or during the processing. PROC_STATUS reports errors found in the telemetry or during the processing. |
|||
| PROC_STATUS[1,2]==b0 means that the data in the row was good and processed correctly. | |||
We recommend that users create a separate directory for analysis, although they can analyze data under the event_cl directory. Hereafter, the analysis directory is called analysis/.
Users should copy or link the following files to analysis/.
| 000126000 /auxil/xa000126000 .ehk |
| 000126000 /resolve/event_cl/xa000126000 rsl_p0px1000_cl.evt |
| 000126000 /resolve/event_uf/xa000126000 rsl_px1000_exp.gti |
| 000126000 /xtend/event_cl/xa000126000 xtd_p030000010_cl.evt |
| 000126000 /xtend/event_uf/xa000126000 xtd_p030000010.bimg |
See Section 4.3 for the file convention. If users run xapipeline, all these files are under the outdir directory.
Users perform further data screening or processing optimized for the targets and then produce images, light curves, and spectra. The procedures are more specific to the instruments, so we describe them separately in the next two chapters. The point and diffuse source analyses share most reduction procedures except for the arf response creation. We explicitly describe their differences when necessary.
Bright sources need extra care as the instrument's behaviors change at high count rates. The flux definition of bright sources is different between Resolve and Xtend instruments or the applied observation mode (See Chapter 8 in POG for Resolve and Section 6.3 in POG for Xtend) and there are multiple count rate thresholds that change the behaviors. We describe the issue briefly in the last section of each chapter, but the detailed treatment is beyond the scope of this document.