THE JUST-THE-BASICS GUIDE TO GETTING STARTED WITH SCISERVER
NASA and Johns Hopkins University have developed SciServer, which allows researchers to reprocess their data with SAS remotely on servers at JHU. People who are interested are strongly encouraged to take their time and examine the tutorials and documentation available on the SciServer site in order to best take advantage of this powerful new system. However, the GOF acknowledges that there are times when all a person really wants is a fast and simple on-ramp. This is intended to be just that: a quick-start guide to using SAS in SciServer.
SciServer now supports visualization using ds9, so software that has graphics, like Sherpa, fv, and Xspec, can be used without worrying if, for instance, a light curve or fitted spectrum can be displayed. However, if users think that lag may be a problem, they are encouraged to have visualization software on their own computers and download the data they are interested in.
Now on with the show!
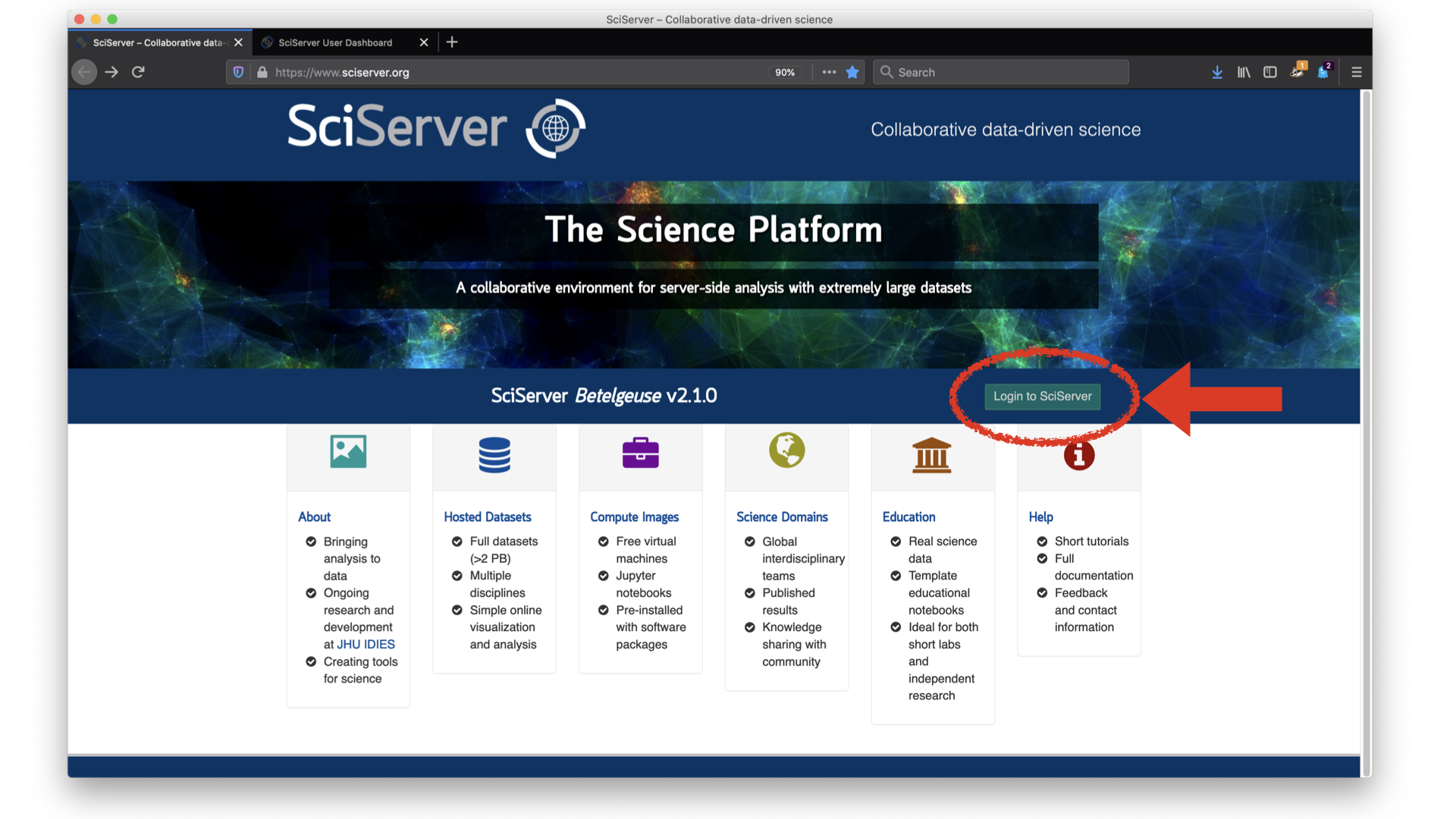
The first thing we will need to do is make an account and log in:
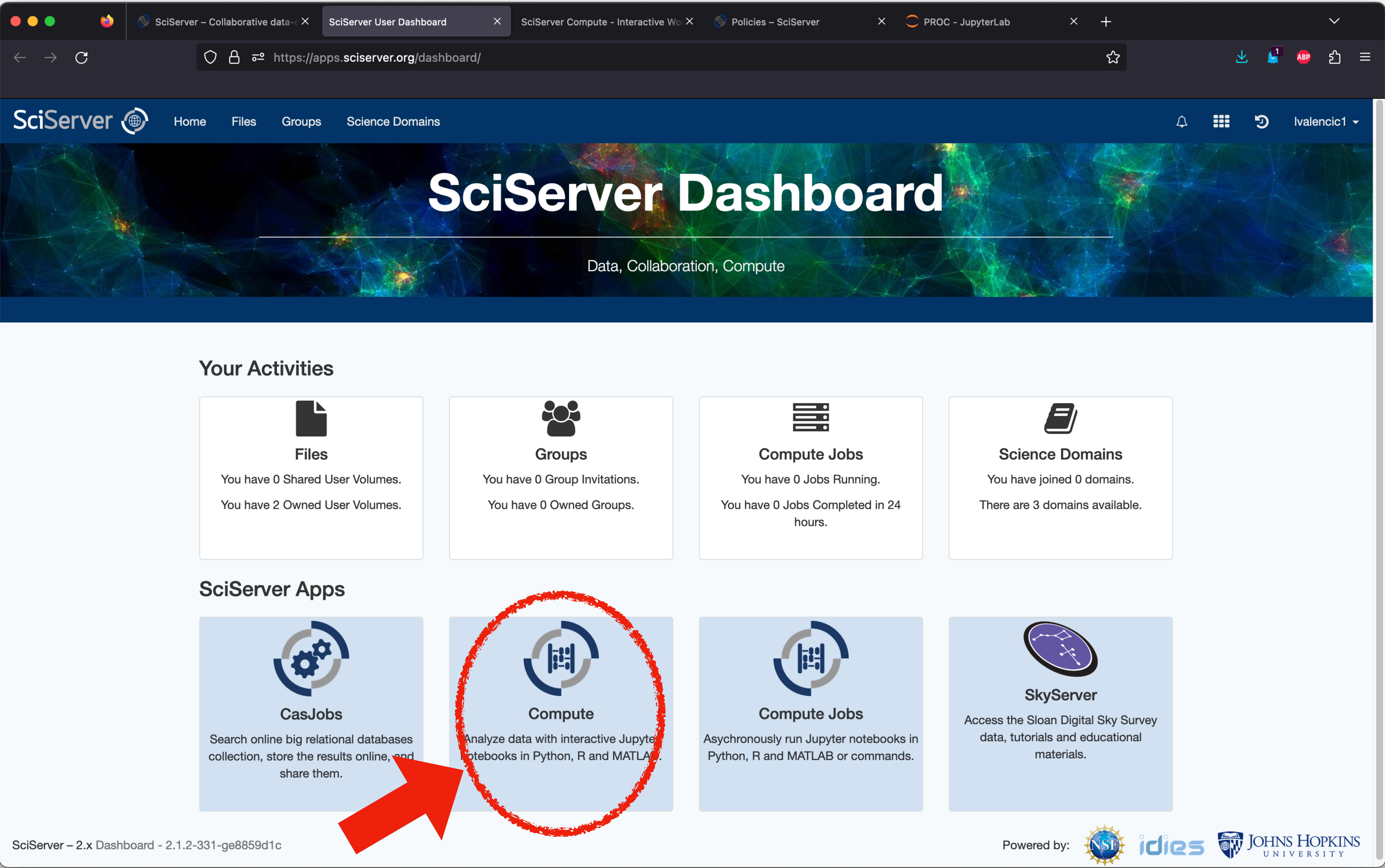
Once we have logged in, a new tab will open that shows the SciServer Dashboard (see Figure 2). On the Dashboard, click on Compute:
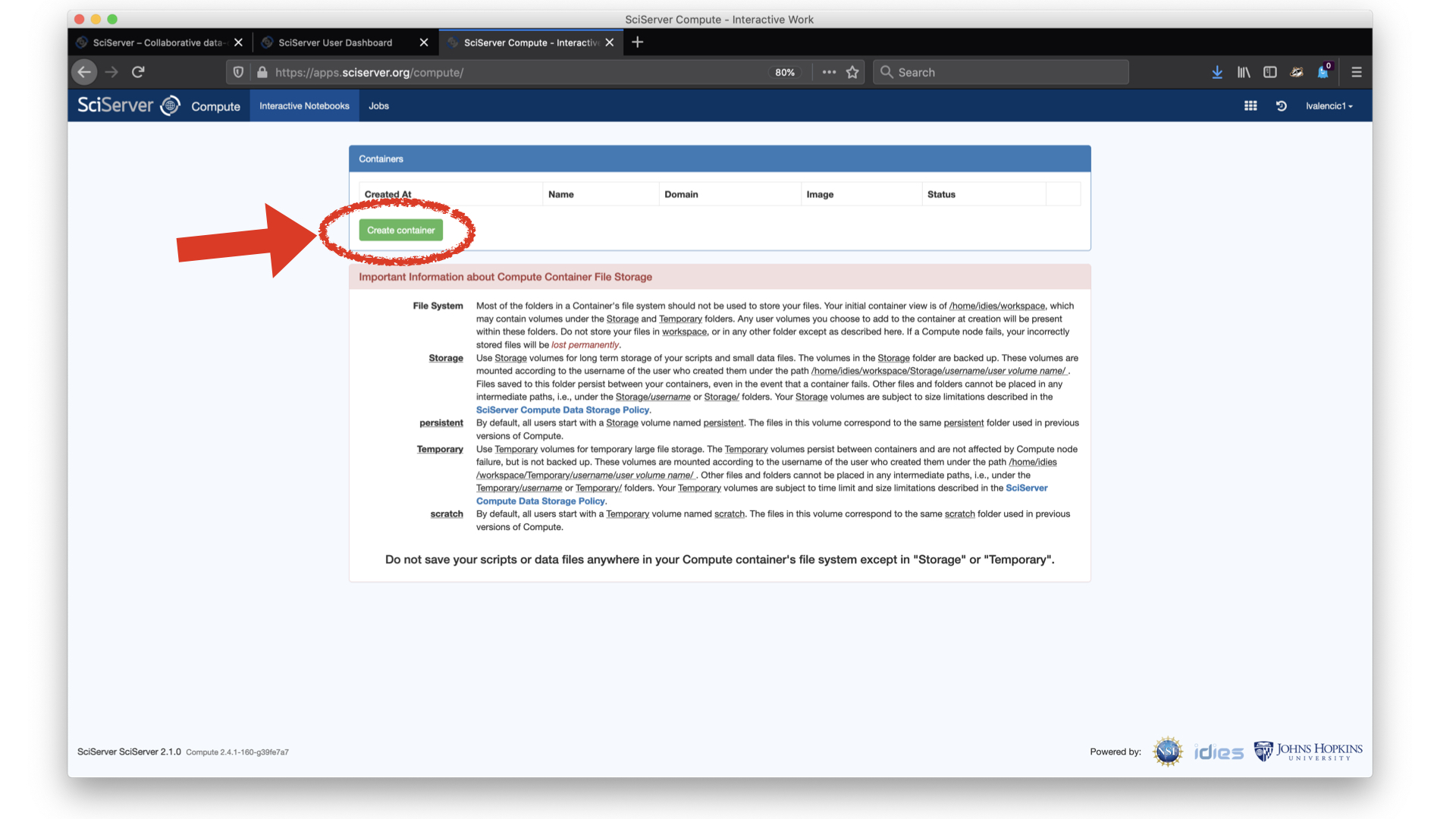
This will open another tab that shows the SciServer Compute - Interactive Work page (see Figure 3), which will give us the opportunity to start a container. Before doing so, it is strongly recommended that users read the information about file storage. There are three main takeaways:
- files stored in the Storage and Temporary directories persist between containers, but only those in Storage are backed up
- the Temporary directory is shared between users. As such, data in Temporary may be deleted after 3 days to maintain sufficient space for all users.
- the Storage directory is 10G per user of permanent space.
With that understanding, we can now click on "Create Container".
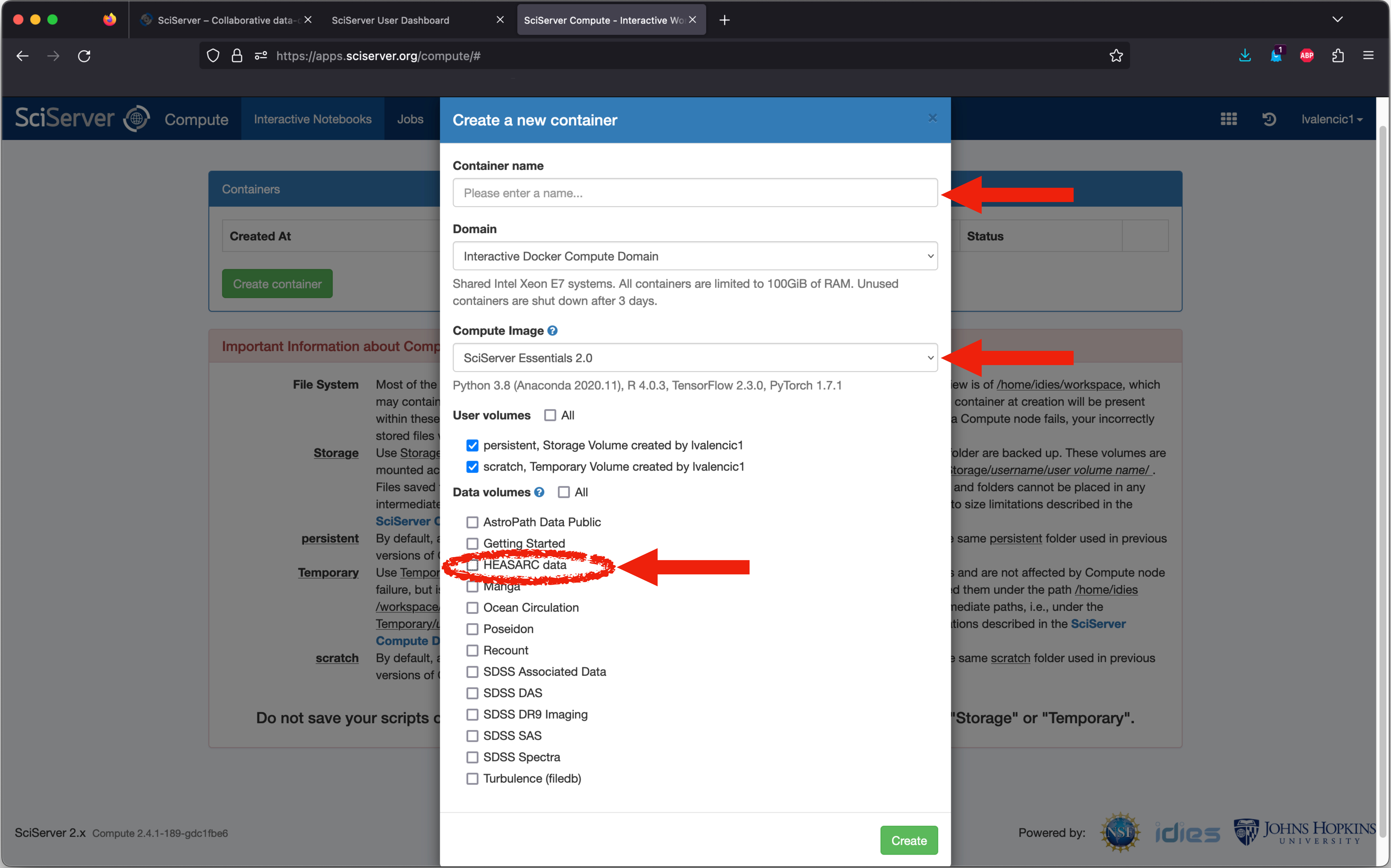
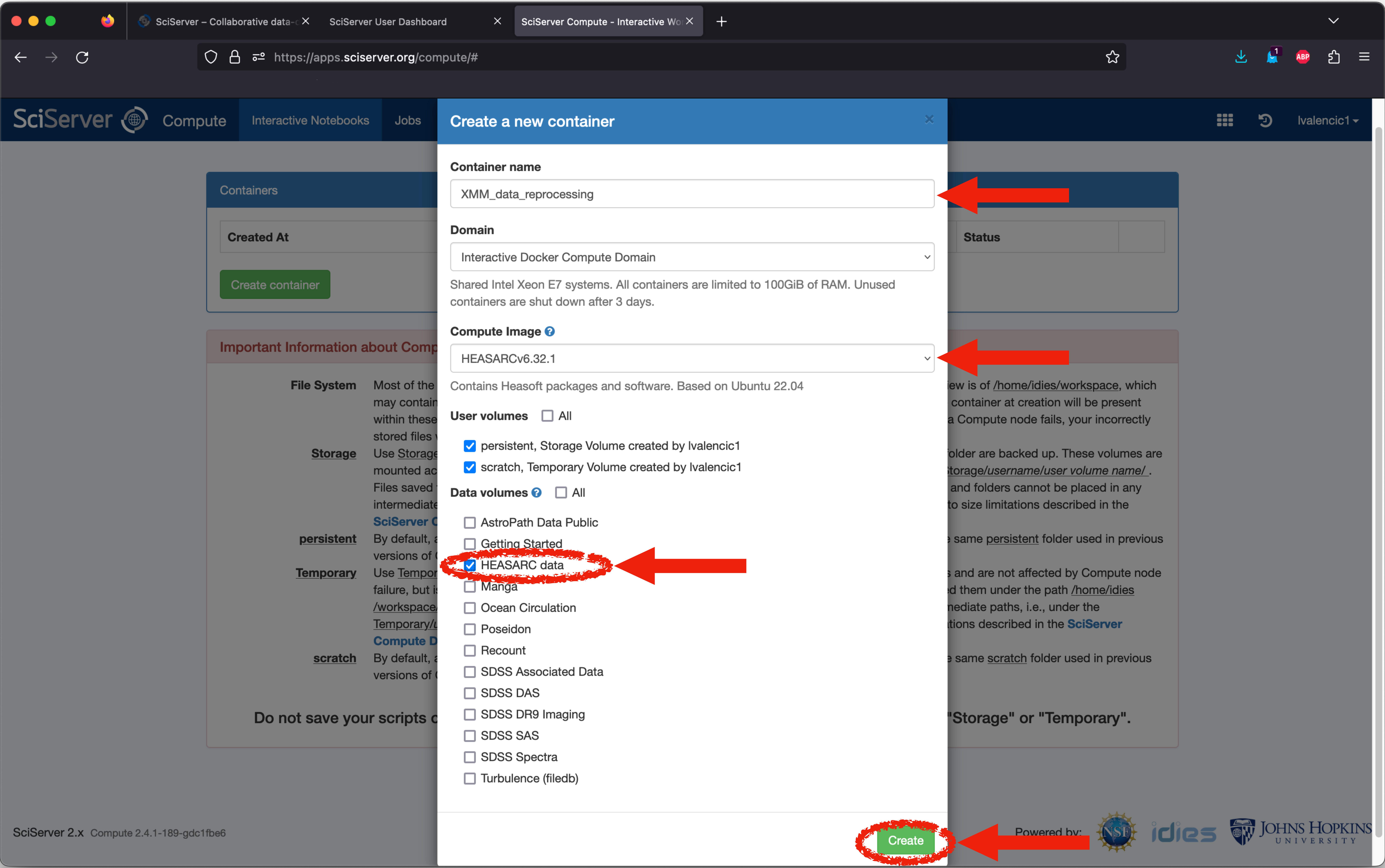
This will open a pop-up window (see Figure 4) where we can name the container and define its attributes in the Compute Image drop-down menu, and User Volumes and Data Volumes selections:
We will need to be able to access SAS in our container. This is done by selecting "HEASARCv6.32.1" in the Compute Image box's drop-down menu. We also want a persistent Storage Volume and scratch Temporary Volume in User Volumes, which are selected by default. Under Data Volumes, we will want "HEASARC data", so we can copy whatever dataset we need from the archive into our working directory. With this set, we can now create the container:
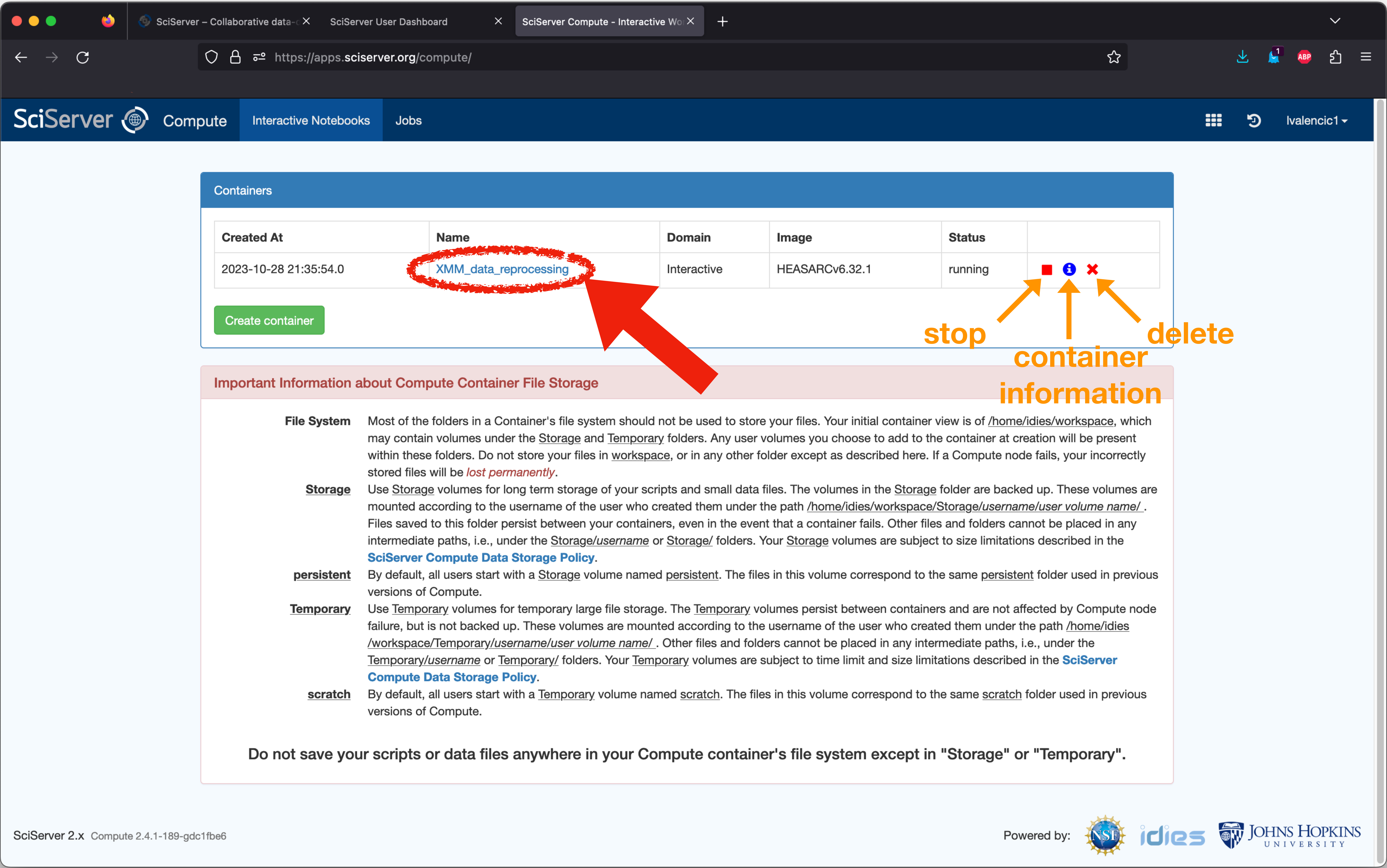
Now in the Interactive Work tab, you will see your container and information about when it was created, its domain, compute image, and status (see Figure 6). There are also small icons that let you stop a running container, get information about it, or delete it.
|
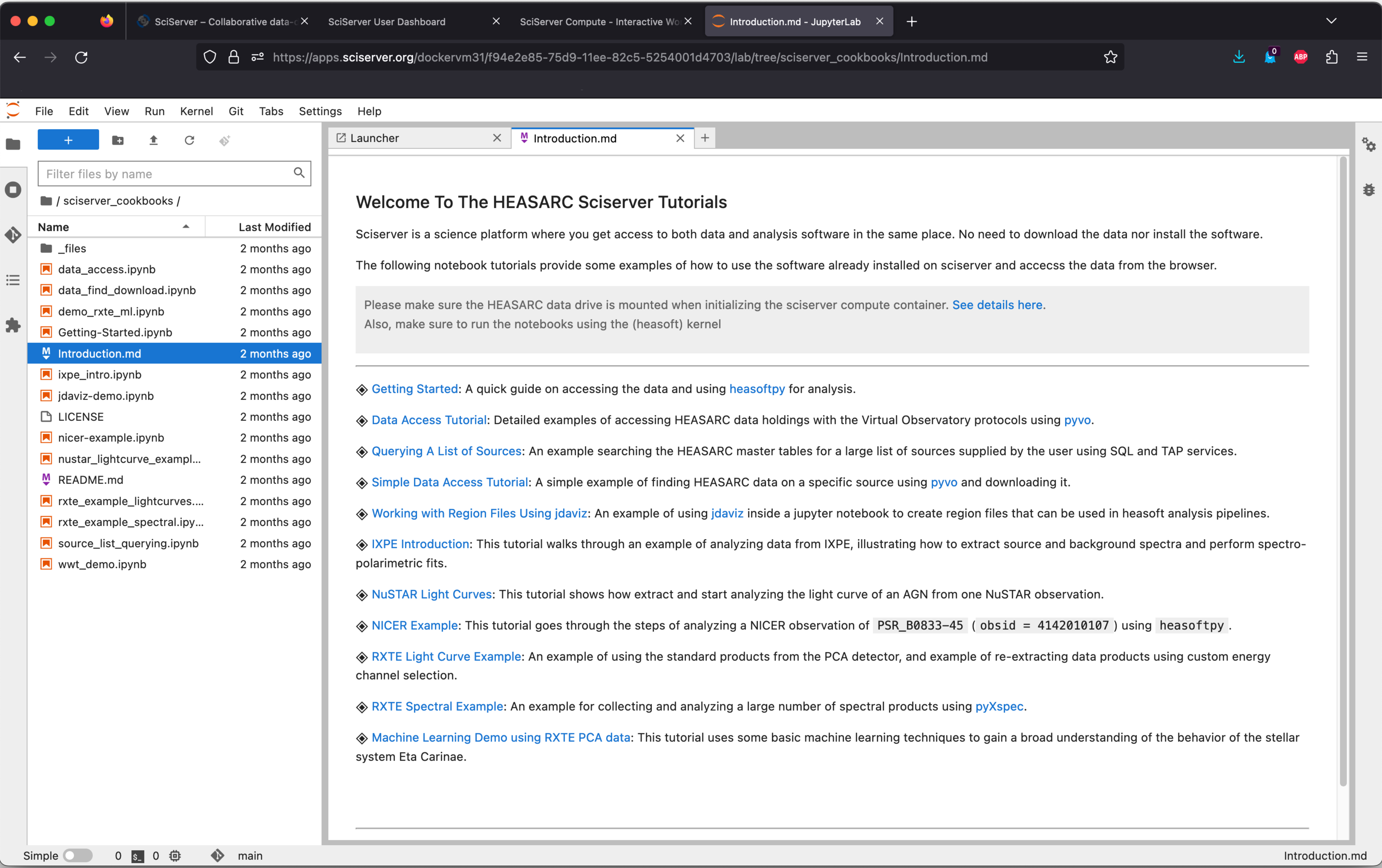
Clicking on the name of the container will open a JupyterLab tab in your browser (see Figure 7) with links to SciServer tutorials in a "minitab" named "Introduction.md". New users are strongly encouraged to read these. Clicking on a link will open a notebook in a new minitab.
To the left of the minitabs is a smaller window with graphical directory navigation. The notebooks which the Introduction.md minitab links to can be seen here, and opened by double-clicking them. Again, a new minitab will be opened for each.
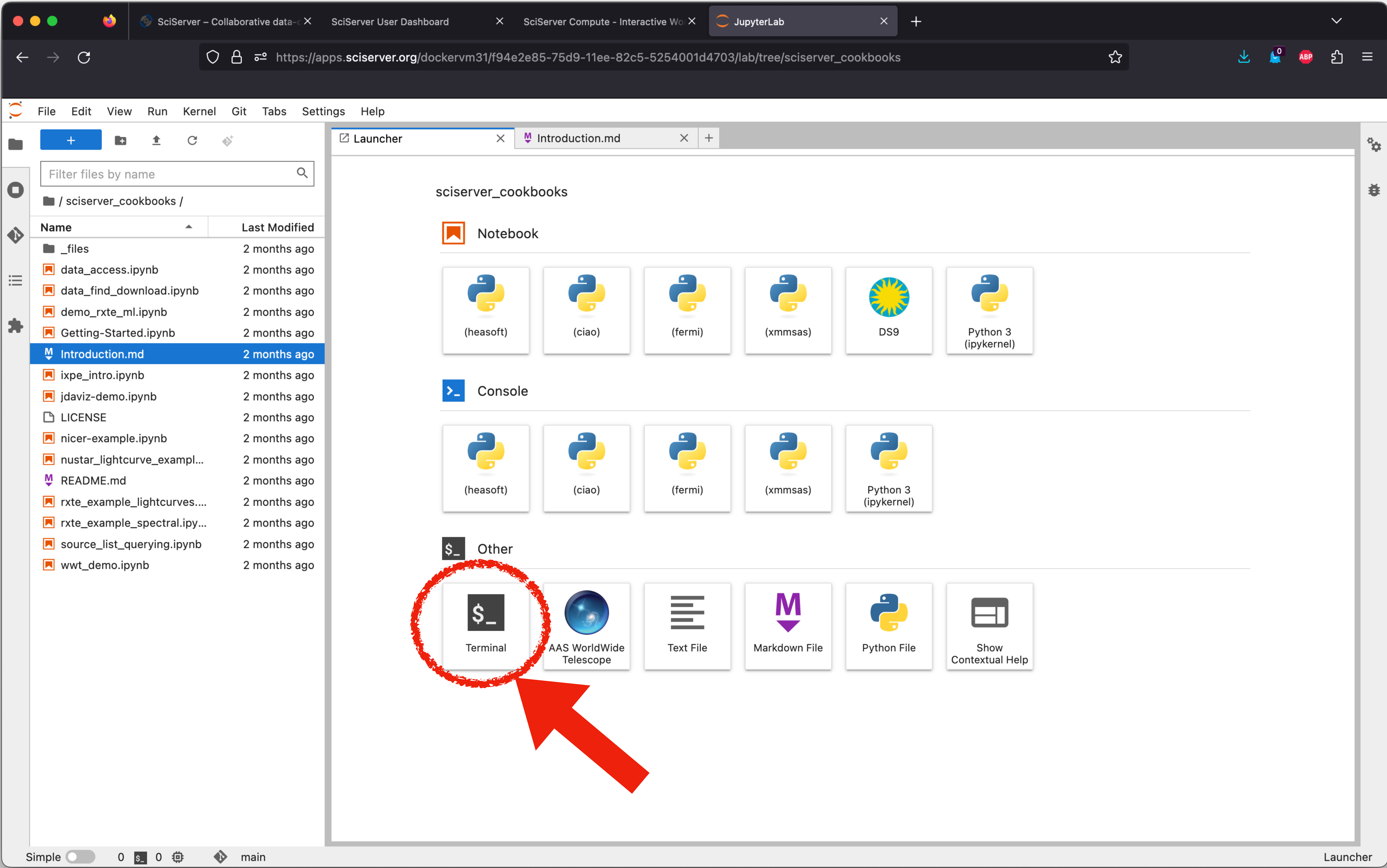
As seen in Figure 7, to the left of the Introduction.md minitab is another minitab called "Launcher". Selecting this tab will display a page of interface options and other actions, like making a text file and opening a notebook (see Figure 8). In the Launcher, in the bottom-most row, select "Terminal".
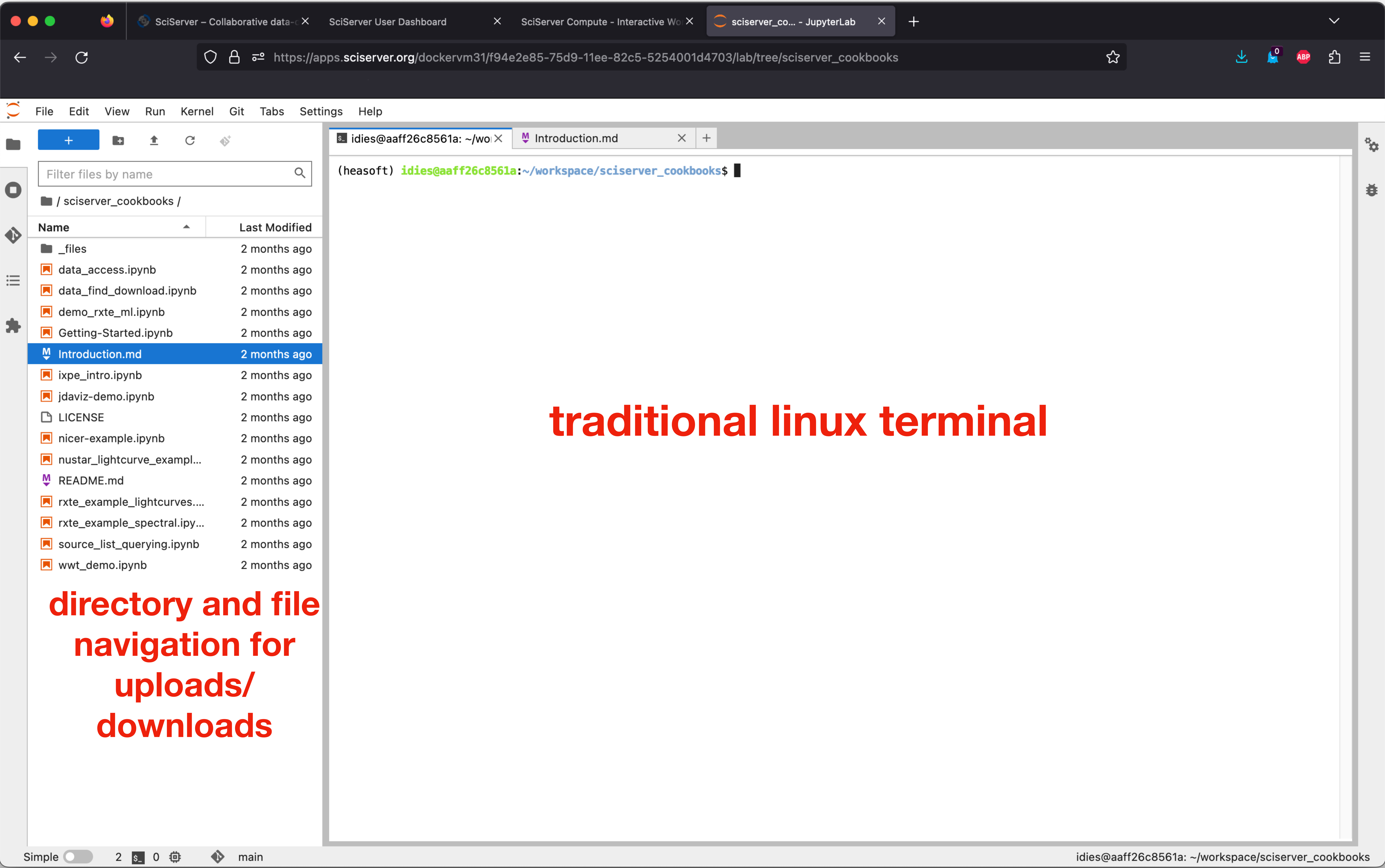
This will open a traditional linux terminal in a new minitab (see Figure 9). Be aware that the linux terminal minitab and the graphical navigation window are not linked, so moving to a different directory in one window does not move us to that directory in the other.
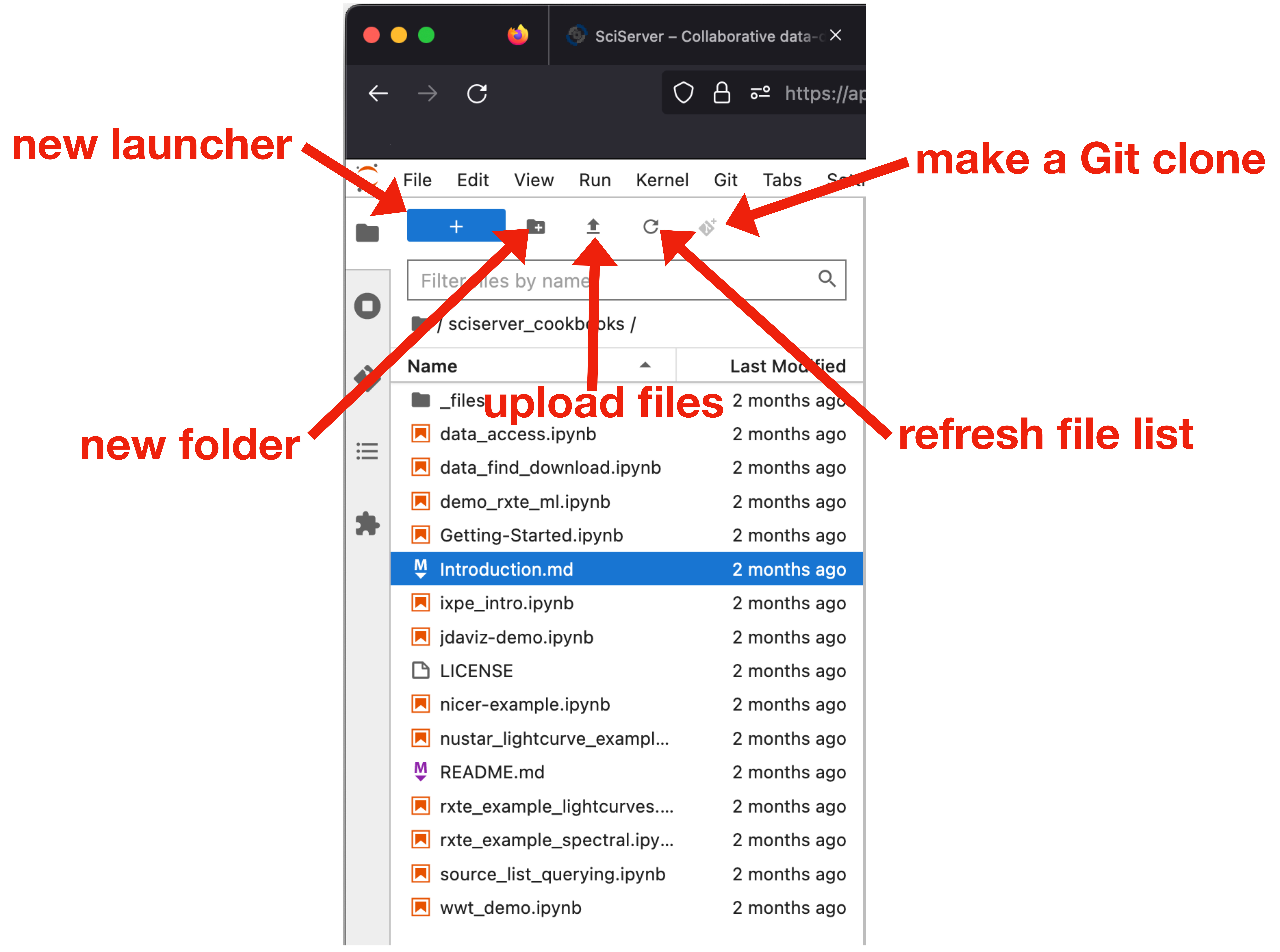
Taking a closer look at the navigation window, above the box that says "Filter files by name", there are five icons (see Figure 10). From left to right, these let us open a new launcher minitab, make a new folder, upload files, refresh the file list, and make a Git clone. In the file list, double-clicking on a folder name will open it; right-clicking on a file will open a menu of options for that file, one of which is to download it.
Now that we have a terminal, we can get some data and initialize SAS.
If we are working on Lockman Hole data (Obs ID 0123700101), we can set up the
workspace and final repository like this, being sure to swap in the correct
username in the commands below:
- mkdir workspace/Temporary/your_username/scratch/LockmanHole
- mkdir workspace/Storage/your_username/persistent/LockmanHole
SciServer has direct access to archived data, and we know which Obs ID we want, so
we can copy it over easily and uncompress it:
- cd workspace/Temporary/your_username/scratch/LockmanHole
- mkdir -p 0123700101/ODF
- cp /home/idies/workspace/headata/FTP/xmm/data/rev0/0123700101/ODF/* 0123700101/ODF
- cd 0123700101/ODF
- gunzip *gz
- mkdir -p 0123700101/ODF
Now we need to initialize SAS and set the standard environment variables:
- source /opt/xmmsas/xmmsas_20210317_1624/setsas.sh
- export SAS_CCFPATH="/FTP/caldb/data/xmm/ccf"
- export SAS_ODF=`pwd`
- export SAS_ODFPATH=`pwd`
- export SAS_CCFPATH="/FTP/caldb/data/xmm/ccf"
And now we can run the SAS setup tasks and reset the environment
variables as usual, in preparation to run the standard processing tasks:
- cifbuild
- export SAS_CCF=`pwd`"/ccf.cif"
- odfingest
- export SAS_ODF=`pwd`'/'`ls *SUM.SAS`
- cd ..
- mkdir PROC
- cd PROC
- export SAS_CCF=`pwd`"/ccf.cif"
At this point, the standard repipelining software can be run and the usual filtering can
be done as shown in the ABC Guide.
Event files, light curves, and images can be viewed using the ds9 app, which can be found
in the Launcher. When we are done, make a tarball of the reprocessed data and move it to Storage:
-
- cd /home/idies/workspace/Temporary/your_username/scratch/LockmanHole/0123700101
- mkdir /home/idies/workspace/Storage/your_username/persistent/LockmanHole/0123700101
- tar cvfz PROC.tgz PROC/*
- mv PROC.tgz /home/idies/workspace/Storage/your_username/persistent/LockmanHole/0123700101
- cd /home/idies/workspace/Temporary/your_username/scratch/LockmanHole/0123700101
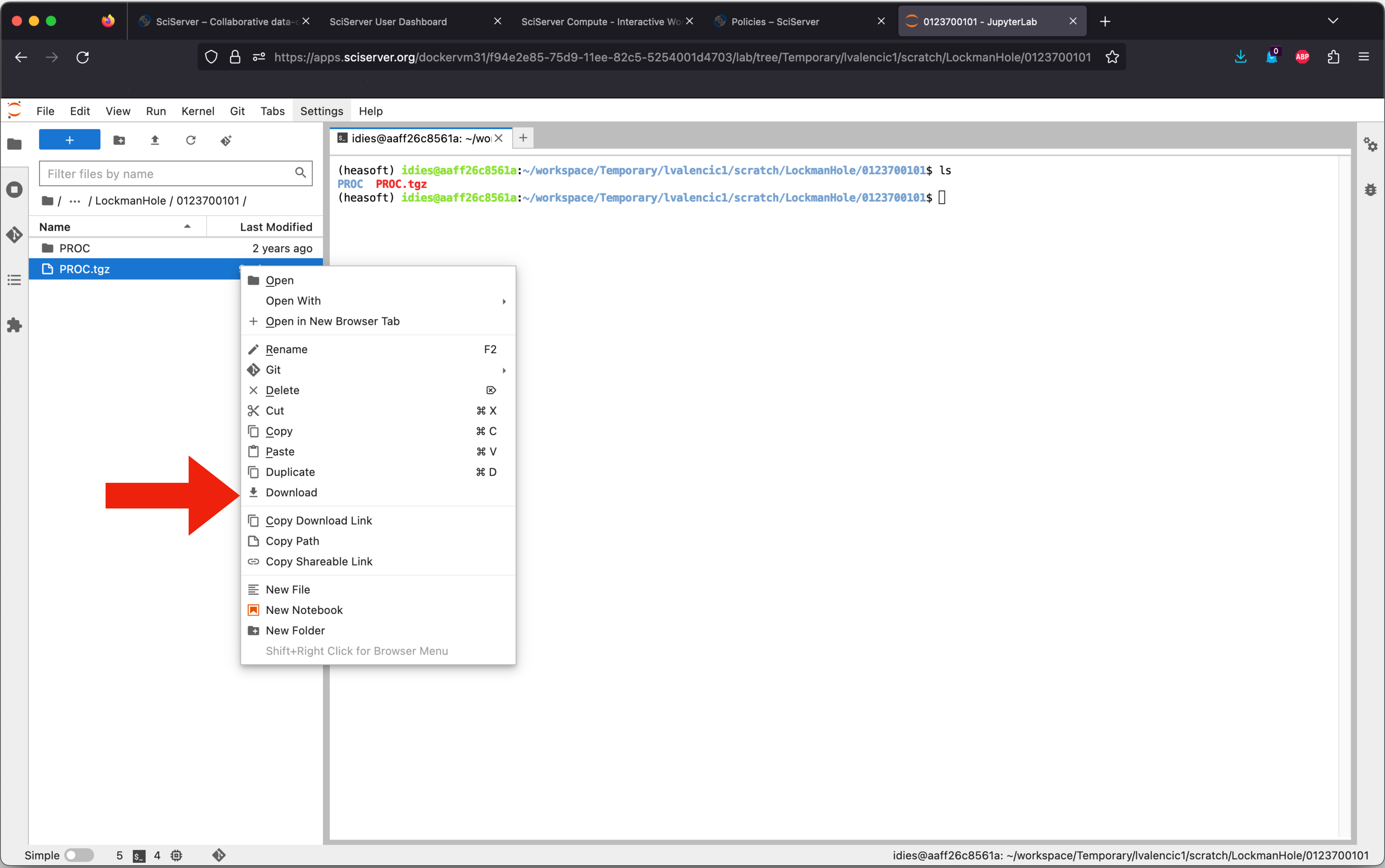
Finally, save it to your local machine by right-clicking on the tarball in the navigation window and selecting "Download" (see Figure 11).
It should be noted that, upon occasion, SciServer gets overloaded. When this happens, users cannot access their containers. In that case, it is recommended that users try again a bit later when the server is not as busy. As a last resort, users might need to remake their containers (i.e., delete their current container and rebuild it from the HEASARC image.) In this case, anything that was not stored in Temporary or Storage is deleted.
If you have any questions concerning XMM-Newton send e-mail to xmmhelp@lists.nasa.gov