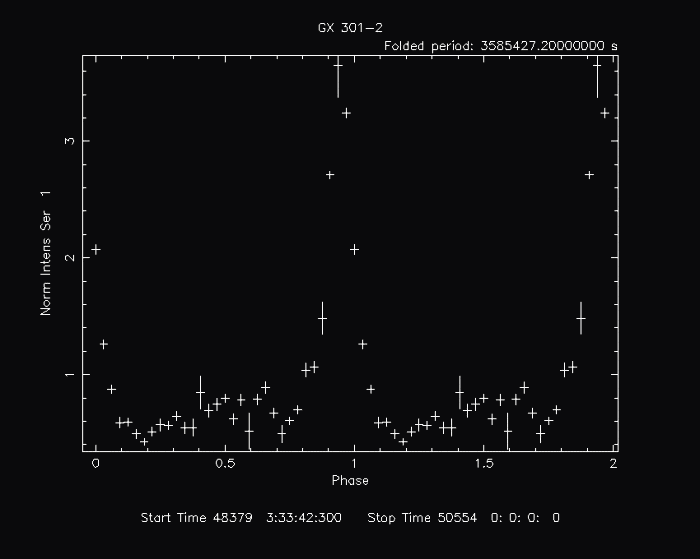
efoldIn this example, we plot the folded light curve of a time series from GRO BATSE of the high mass x-ray binary, GX 301-2. The light curve data is contained in batse_981.lc:
Executing the efold task (Screen output is in a fixed width font, while commentary is italicized): > efold perfo=1 Note that efold is executed with the hidden parameter perfo set to 1, indicating that the period is given in days rather than the default in seconds (perfo=2).
efold 1.1 (xronos5.18) Number of time series for this task[1] Ser. 1 filename +options (or @file of filenames +options)[file1] batse_981.lc We enter the light curve, batse_981.lc directly as the single time series to be considered.
Series 1 file 1:batse_981.lc
WARNING: Defaulting to first FITS extension
Selected FITS extensions: 1 - RATE TABLE;
Source ............ GX 301-2 Start Time (d) .... 48379 00:00:00.000
FITS Extension .... 1 - ` ` Stop Time (d) ..... 50556 00:00:00.000
No. of Rows ....... 1685 Bin Time (s) ...... 0.8640E+05
Right Ascension ... Internal time sys.. Literal
Declination ....... Experiment ........ CGRO BATSE
Corrections applied: Vignetting - No ; Deadtime - No ; Bkgd - No ; Clock - No
Selected Columns: 1- Time; 3- Y-axis; 4- Y-error;
File contains binned data.
Name of the window file ('-' for default window)[-]
Entering a - indicates that the default exposure windows are to be applied ($XRDEFAULTS/default_win.wi). A custom windows file may be generated with the xronwin task.
Expected Start ... 48379.00000000000 (days) 0: 0: 0: 0 (h:m:s:ms) Expected Stop .... 50556.00000000000 (days) 0: 0: 0: 0 (h:m:s:ms) Default Epoch is: 48379.00000 Type INDEF to accept the default value Epoch format is days. Epoch[34 234.23] 48802.79 Entering INDEF would accept the start time of the time series as the Epoch, however, in this example we choose 48802.79 as the Epoch.
Period is in days. Period[88.87] 41.498 As efold was executed with the argument perfo=1, period is assumed in days. Otherwise, the default unit is seconds. In this example, we use a period of 41.498 days.
Expected Cycles .. 52.46
Default phase bins per period are: 10
Type INDEF to accept the default value
Phasebins/Period {value or neg. power of 2}[-3] 32
The phasebins/period (nphase) setting represents the resolution of the final pulse profile (i.e. the number of bins that the range from phase of 0.0 to phase of 1.0 is divided into). If a negative value is given, the phasebins/period is 2 to the power of the absolute value of that number. (e.g. nphase=-3 -> nphase=2abs(-3)=8). The default (10), corresponding to INDEF may be altered with the hidden parameter nbdf.
Newbin Time ...... 112044.60 (s) Maximum Newbin No. 1679 Default Newbins per Interval are: 1679 (giving 1 Interval of 1679 Newbins) Type INDEF to accept the default value Number of Newbins/Interval[10] 1700 Entering INDEF would include exactly the timespan of the input in the resulting plot. Giving a smaller number than the indicated INDEF value would result in multiple intervals. In this example, we use the value 1700.
Maximum of 1 Intvs. with 1700 Newbins of 112045. (s) Default intervals per frame are: 1 Type INDEF to accept the default value Number of Intervals/Frame[1] If multiple intervals exist, frames may be calculated from the average of some of them and then plotted. One frame corresponds to one plot. In this example, we have only one interval, and consequently one frame and one plot.
Results from up to 1 Intvs. will be averaged in a Frame Name of output file[default] In this example, we enter a space before pressing Return to indicate that an output FITS file is not desired, however, entering default for the output file, will result in a folded FITS light curve file to be written with the extension .fef.
Do you want to plot your results?[yes] In general, we want to plot the folded light curve, however, entering no will prevent efold from plotting the output. This is useful when only the output FITS file is desired.
Enter PGPLOT device[/XW] The Xwindows device, /xw is recommended as the results may be viewed immediately and the plot may be manipulated interactively with PLT. In some cases, however, other file-based devices such as /ps and /gif may be desired.
32 analysis results per interval
WARNING: Defaulting to first FITS extension
Intv 1 Start 48379 3:33:42
Ser.1 Avg 0.4701E-01 Chisq 3385. Var 0.1372E-02 Newbs. 32
Min 0.1984E-01 Max 0.1717 expVar 0.1386E-04 Bins 1685
Folded light curve ready
By default, the following plot is produced:
It may, however, by manipulated with any PLT command.
PLT> line step Connect the points.
PLT> color 5 on 2 Change group 2 (the data representing Y) to color 5 (light blue).
PLT> error off Turn error bars off
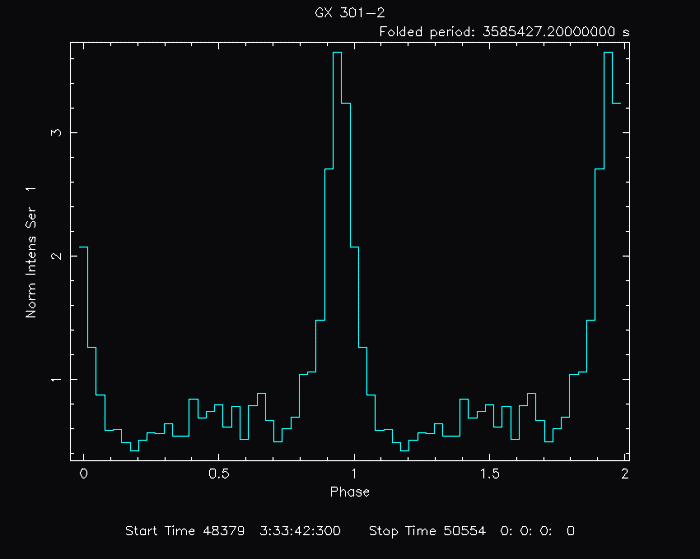
PLT> cpd gx301-2.gif/gif Change the plot device to a gif.
PLT> p Plot to the gif.
PLT> quit The resulting gif file looks like this:
Please send reports of errors to : xanprob@athena.gsfc.nasa.gov HEASARC Home | Observatories | Archive | Calibration | Software | Tools | Students/Teachers/Public Last modified: Friday, 26-Mar-2004 16:35:02 EST |