Spectropolarimetry ExampleXSPEC can fit spectropolarimetric models to spectra for the I, Q, and U Stokes parameters. The following is a toy example for IXPE data. The responses are notional and should not be used with actual data.
XSPEC12> data 1:1 toy_point_source_du1_pha1.fits XSPEC12> data 1:2 toy_point_source_du1_pha1q.fits XSPEC12> data 1:3 toy_point_source_du1_pha1u.fits XSPEC12> data 2:4 toy_point_source_du2_pha1.fits XSPEC12> data 2:5 toy_point_source_du2_pha1u.fits XSPEC12> data 2:6 toy_point_source_du2_pha1q.fits XSPEC12> ignore 1-6:0.0-2.0 8.0-** This has read in the I, Q, U triplet for two of the three telescopes and placed each triplet in its own data group. The I, Q, and U spectra are distinguished by having different values of their XFLT0001 keyword: 'Stokes:0', 'Stokes:1', and 'Stokes:2' for I, Q, and U respectively.
XSPEC12>model polconst*powerlaw
...
========================================================================
Model polconst<1>*powerlaw<2> Source No.: 1 Active/On
Model Model Component Parameter Unit Value
par comp
Data group: 1
1 1 polconst A 1.00000 +/- 0.0
2 1 polconst psi deg 45.0000 +/- 0.0
3 2 powerlaw PhoIndex 1.00000 +/- 0.0
4 2 powerlaw norm 1.00000 +/- 0.0
Data group: 2
5 1 polconst A 1.00000 = p1
6 1 polconst psi deg 45.0000 = p2
7 2 powerlaw PhoIndex 1.00000 = p3
8 2 powerlaw norm 1.00000 = p4
________________________________________________________________________
defines a power-law model with constant polarization fraction and angle.
XSPEC12> fit
...
========================================================================
Model polconst<1>*powerlaw<2> Source No.: 1 Active/On
Model Model Component Parameter Unit Value
par comp
Data group: 1
1 1 polconst A 0.498573 +/- 1.12218E-03
2 1 polconst psi deg 29.9835 +/- 6.40758E-02
3 2 powerlaw PhoIndex 1.99757 +/- 9.75912E-04
4 2 powerlaw norm 9.96930 +/- 1.13268E-02
Data group: 2
5 1 polconst A 0.49857 = p1
6 1 polconst psi deg 29.983 = p2
7 2 powerlaw PhoIndex 1.99757 = p3
8 2 powerlaw norm 9.96930 = p4
________________________________________________________________________
Fit statistic : Chi-Squared 175.42 using 149 bins.
Chi-Squared 107.92 using 149 bins.
Chi-Squared 165.08 using 149 bins.
Chi-Squared 168.22 using 149 bins.
Chi-Squared 133.51 using 149 bins.
Chi-Squared 144.68 using 149 bins.
Total fit statistic 894.82 with 890 d.o.f.
Test statistic : Chi-Squared 894.82 using 894 bins.
Null hypothesis probability of 4.48e-01 with 890 degrees of freedom
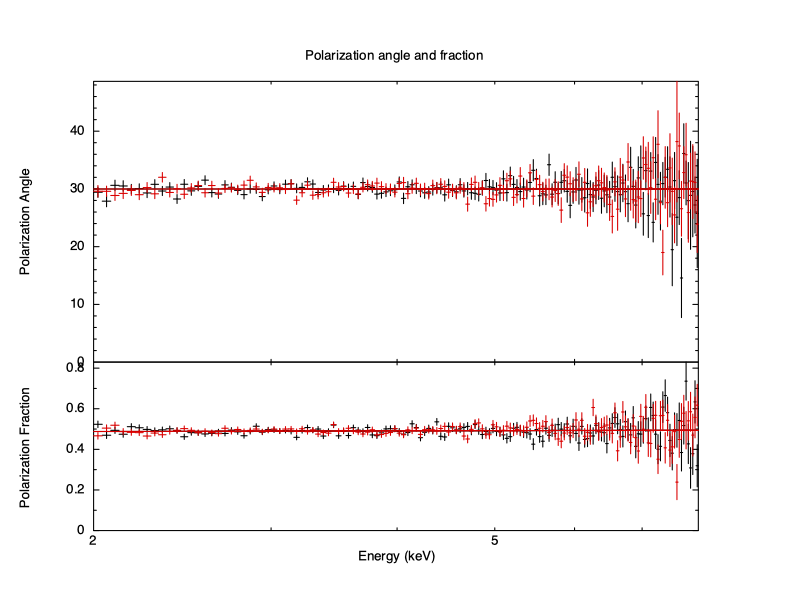
Plotting the data shows the data and model for each individual spectrum as usual but for spectropolarimetry there is an additional plot option to show the derived polarization angle and fraction for both the data and the model as calculated from the Stokes parameters.
XSPEC12> plot polangle polfrac
HEASARC Home | Observatories | Archive | Calibration | Software | Tools | Students/Teachers/Public Last modified: Friday, 23-Aug-2024 13:20:40 EDT |