Resolve NXB Database and Spectral Extraction Recipes
Overview
This page is to share the "provisional" NXB Database (DB) of Resolve. This page also provides an example of how to run rslnxbgen with the database under HEASOFT 6.34. For this provisional DB (and your source), we recommend users to select only Hi-res events and apply screening (shown below) manually.
Data Files
- NXBDB
- Science data for your source; here we use OBSID 123456789 as an example. Replace with your data OBSID in the commands below.
- Data event file: xa123456789rsl_p0px1000_cl.evt
- ehk file: xa123456789.ehk
- Please replace with your own data files via commands below
- Other files
- Repository
Example of Commands
Prepare "base" GTI for source and NXB (T_SAA_SXS>0 is equivalent to SAA_SXS==0) to make them compatible with each other. CORTIME cut for source is optional and you shall apply the same cut to NXB in running rslnxbgen ("sortbin" option)
- maketime merged_reduced_rev3_fix2.ehk ehkSelA.gti "T_SAA_SXS>0 && ELV<-5 && DYE_ELV>5" compact=no time=TIME no
- maketime xa123456789.ehk.gz ehkSelB.gti "T_SAA_SXS>0 && DYE_ELV>5 && CORTIME>6" compact=no time=TIME no
GTI-based selection on the NXB event file.
- extractor 'filename=merged_nxb_resolve_gtifix.evt' 'eventsout=merged_nxb_resolve_gtifix_ehkSel.evt' 'imgfile=NONE' 'phafile=NONE' 'fitsbinlc=NONE' 'regionfile=NONE' 'timefile=ehkSelA.gti' 'xcolf=X' 'ycolf=Y' 'tcol=TIME' 'ecol=PI' 'xcolh=DETX' 'ycolh=DETY'
- ## 87871, 784.9ks
GTI-based selection on the source file.
- extractor 'filename=xa123456789rsl_p0px1000_cl.evt.gz' 'eventsout=xa123456789rsl_p0px1000_cl_ehkSel.evt' 'imgfile=NONE' 'phafile=NONE' 'fitsbinlc=NONE' 'regionfile=NONE' 'timefile=ehkSelB.gti' 'xcolf=X' 'ycolf=Y' 'tcol=TIME' 'ecol=PI' 'xcolh=DETX' 'ycolh=DETY'
Event-based selection on the source event file.
- ftselect 'xa123456789rsl_p0px1000_cl_ehkSel.evt[EVENTS]' xa123456789rsl_p0px1000_cl_ehkSel_evSel.evt "(PI>=600) && ((RISE_TIME+0.00075*DERIV_MAX)>46) && ((RISE_TIME+0.00075*DERIV_MAX)<58) && (ITYPE==0) && (STATUS[4]==b0) && (PIXEL!=27)"
Run rslnxbgen to extract NXB spectrum for the entire array (except PIXEL=12&27). Also, you may adopt larger values for timefirst and timelast if your data was taken well after this NXBDB (accumulates data from Dec. 25, 2023 to May 24, 2024).
- rslnxbgen infile=xa123456789rsl_p0px1000_cl_ehkSel_evSel.evt ehkfile=xa123456789.ehk.gz regfile=NONE pixels=- innxbfile=merged_nxb_resolve_gtifix_ehkSel.evt innxbehk=merged_reduced_rev3_fix2.ehk database=LOCAL db_location=./ timefirst=-150 timelast=+150 SORTCOL=CORTIME sortbin="6,8,10,12,99" expr="(PI>=600) && ((RISE_TIME+0.00075*DERIV_MAX)>46) && ((RISE_TIME+0.00075*DERIV_MAX)<58) && (ITYPE==0) && (STATUS[4]==b0) && (PIXEL!=27)" outpifile=rslnxb.pi outnxbfile=rslnxb.evt outnxbehk=rslnxb.ehk
Extract source spectrum from the entire array (except PIXEL=12&27)
- #-----
- xselect
- XRISM
- read events
- ./
- xa123456789rsl_p0px1000_cl_ehkSel_evSel.evt
- yes
- extract events
- extract spectrum
- save spectrum
- src.pi
- set image det
- extract image
- save image
- src_det.img
- exit
- no
- #-----
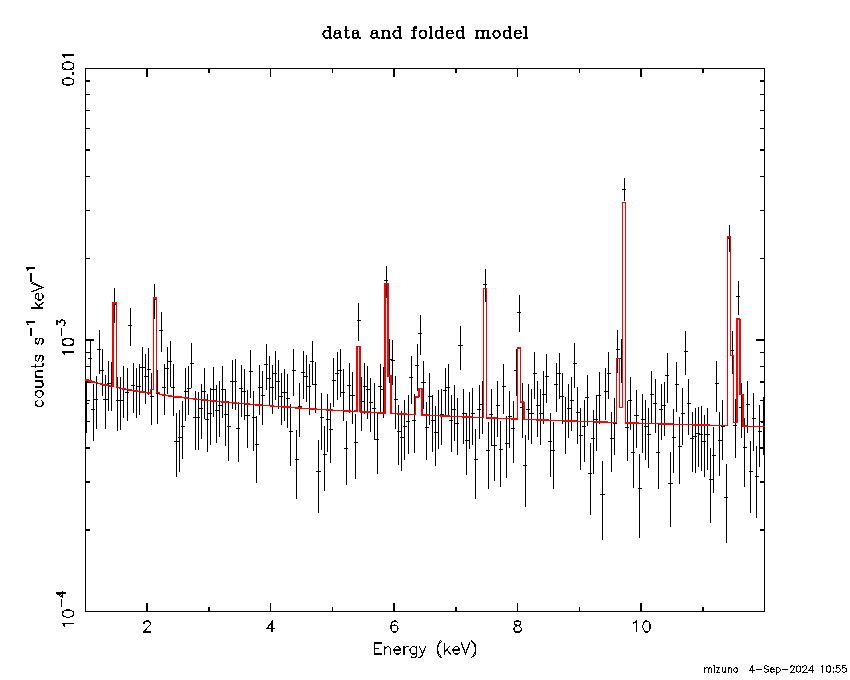
Generated NXB Spectrum vs. Model
You may check the generated NXB spectrum as shown below. Normalizations for continuum and lines other than Mn K-alpha are tied together, and the nomalization of Mn K-alpha is adjusted independently. Link to xcm file.
- #-----
- xspec
- data 1:1 rslnxb.pi
- resp 1:1 newdiag60000.rmf
- setplot device /xw
- setplot energy
- ig **-1.0 12.0-**
- plot
- setplot rebin 1000 100
- plot
- @test-rslnxbgen-nxbmodel.xcm
- show
- newpar det:1 0.9
- newpar det:4 1.0
- save all test-rslnxbgen-nxbmodel.xcm
- setplot xlog off
- iplot 1d
- rescale y 1e-4 1e-2
- color 2 on 2
- plot
- hard /gif
- exit
- exit
- #-----
- mv pgplot.gif test-rslnxbgen-nxbmodel_plot.gif