HEASARC@SciServer User GuideCurrent build "HEASARCv6.31.1": HEASoft v6.31.1, Ciao 4.15, XMM SAS v20, and Fermitools 2.2.0.Here's an eleven minute video tutorial
Or click here to download.
Contents
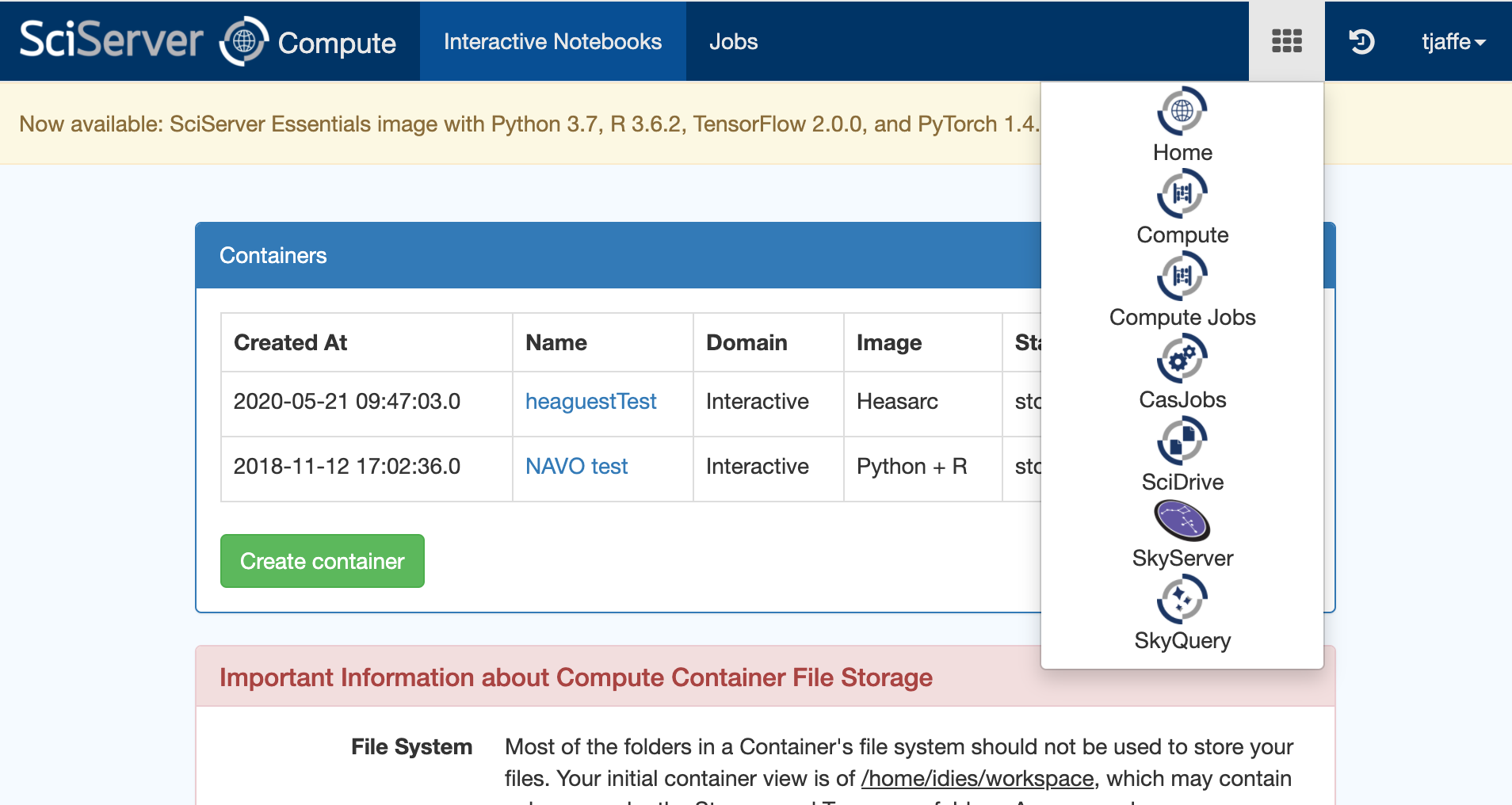
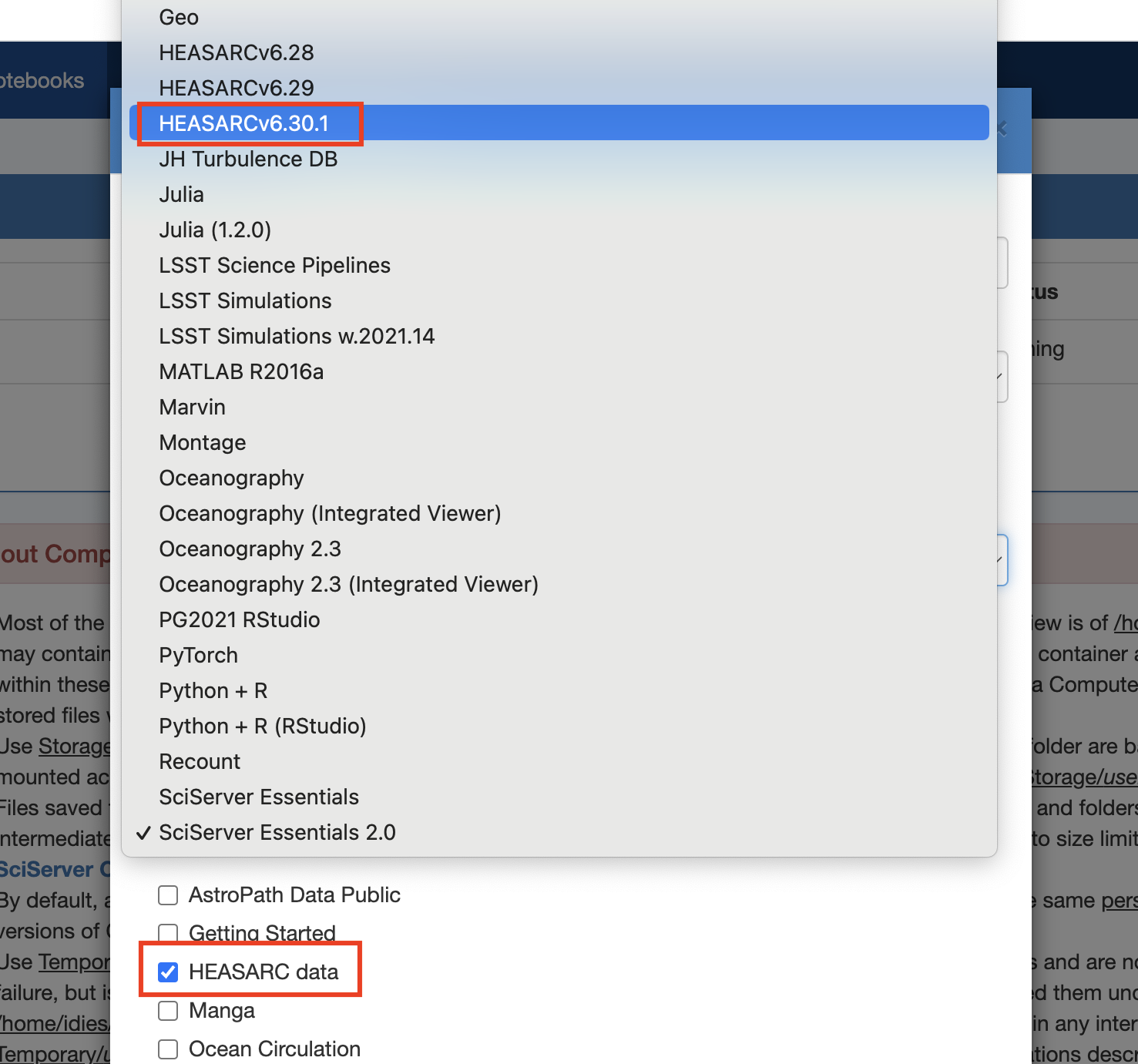
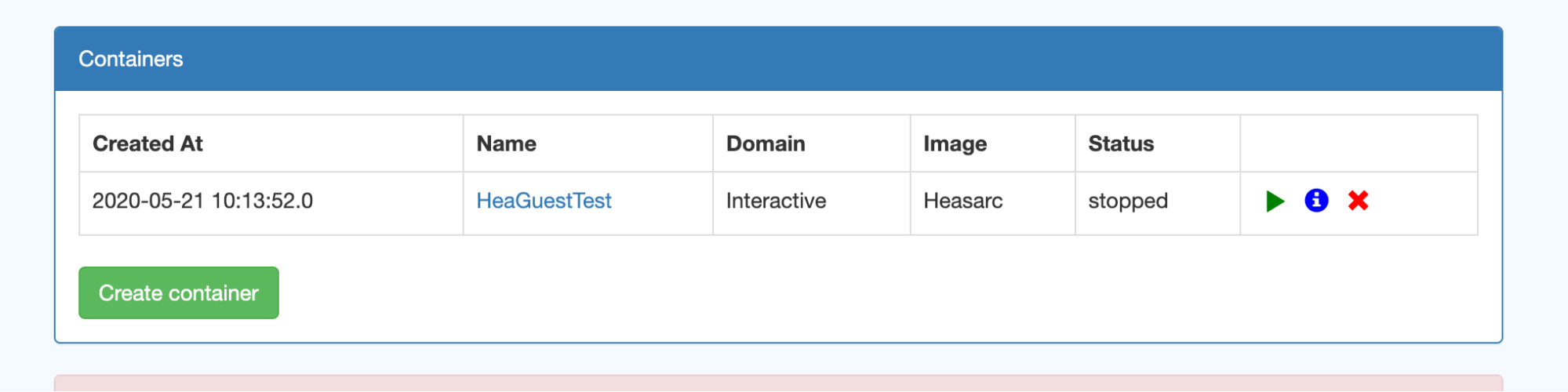
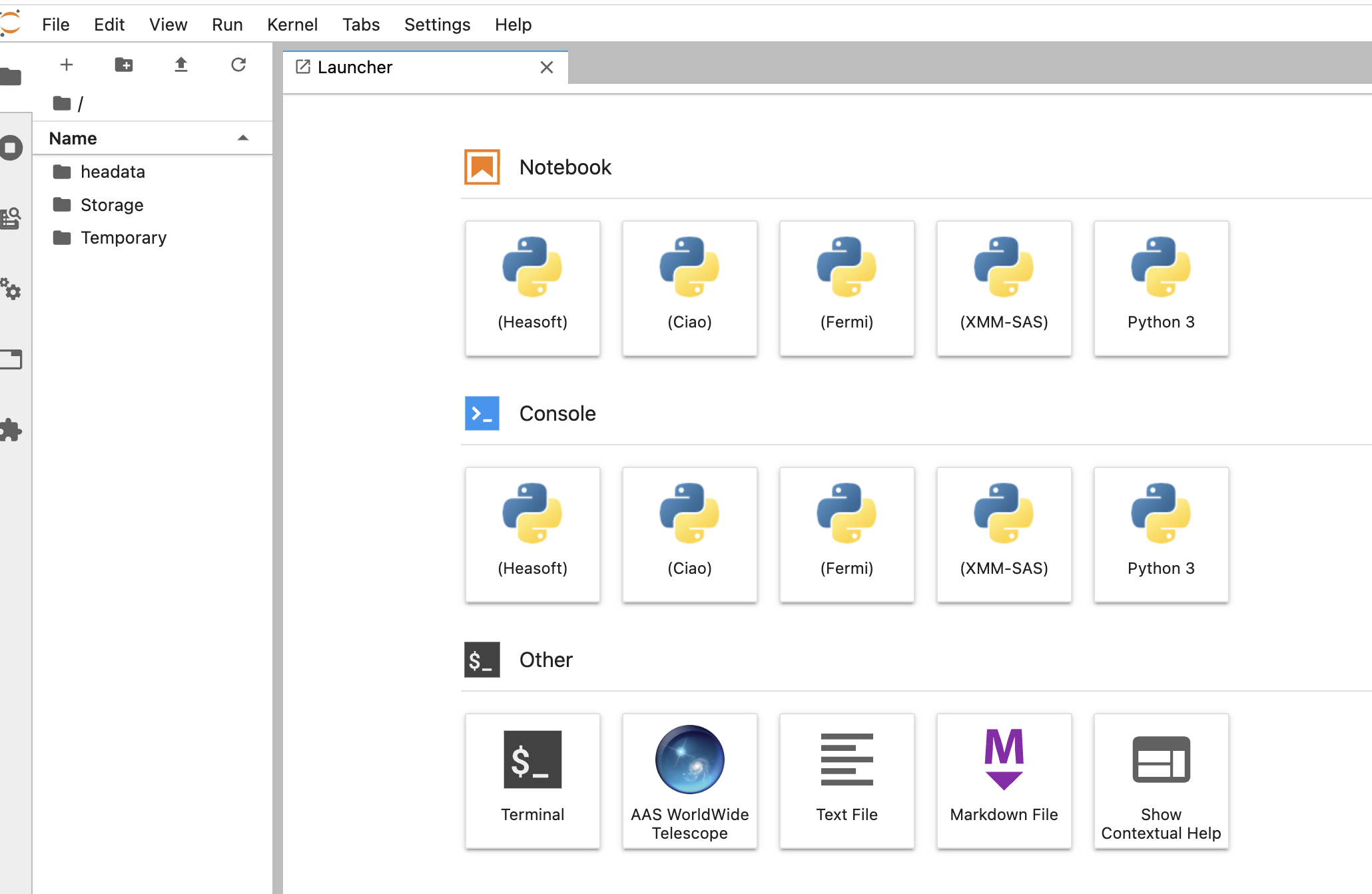
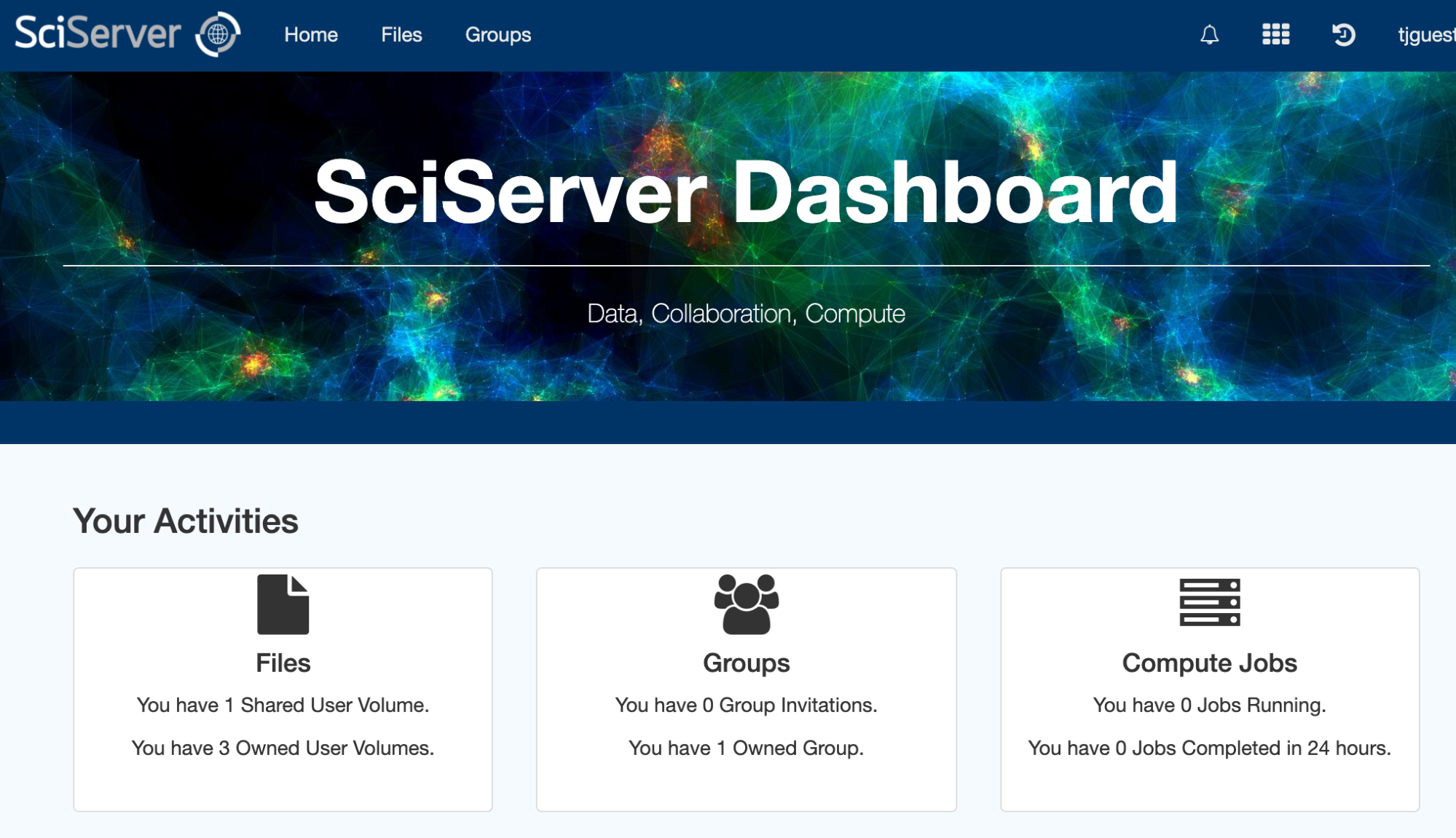
If you encounter any issues running code or accessing the data, please contact the HEASARC help desk. If your issue is not related to the HEASARC setup specifically, then you can email the SciServer helpdesk. Click on your username at the top right of your dashboard and select Help from the dropdown menu.I. Getting startedCreate your own account on https://www.sciserver.org/. Then go to "SciServer -> Compute" (note: not "Compute Jobs", which is the batch). It will list your containers, and if this is the first time you've looked, there won't be any. Here's a screen shot: Note the grid symbol on the top menu bar that allows you to get around. The Compute is the second down. The first is the Home dashboard, where you'll find your files and groups, which we'll get to later. Click "Create container". You'll be presented with some options. First, enter a name for your container. (This can be done repeatedly, so don't worry too much.) Then ignore the Domain. Third is the Compute Image your container will be based on. Select HEASARCv6.30.1 (or other versions) from the drop-down menu. Then further down, your User Volumes are shown (more later). Below that are the Data Volumes where you should select the HEASARC data volume. This will make our data and software areas accessible from within your container. When you've created it, you'll see its status in the list. In this screen shot, it isn't already running, so there's a green arrow to start it. The red x lets you delete it. If the green arrow is instead a red square, that means it's already running, and you can stop it if you wish. Note that if you close your browser without stopping the container, it'll still be running in the same state in which you left it. If you click on the name of the container, a new tab will open with a Jupyter Lab interface. There are 4 conda environments for each of the provided packages. The default environment is (heasoft) and is configured to run Heasoft and Heasoftpy: From here, you can either start a Jupyter Notebook, a Console, or just run a Terminal. You can also navigate to one of our Jupyter notebooks (more below) or upload your own and run it here.II. File systemsThere are a couple of things to know about the file systems available to you within the container. By default, you'll have areas called "temporary" and "persistent", which are what they sound. The persistent area is going to have the same contents if you stop the container and go back to it later and will be visible from other containers you define. It has a limit of 10GB total. These will be under your HOME directory:
which contains
If you choose to define your own user volume (see below) and mount it to this container, it will also appear here, as will any that are shared with you by other users under the corresponding username:
So under Storage is where you should put your work if you want it to survive outside the containers. If you want to share stuff, you should create a user volume rather than stash it in persistent, which nobody else can see. You should also back it up yourself, see Miscellaneous below. If you chose to mount the HEASARC data volume (when creating the container), it will be found at the same level as your Storage and Temporary areas, eg., you'll see
The FTP area will contain all of the HEASARC data holdings exactly as they are organized on our own FTP site. Our compute image also puts a link to this area at /FTP (though this link will be broken if you forget to mount the volume). The default software installation is not on that volume but built within the compute container under
The image is non-trivial to update but will be kept up to date with major software releases. The data volume, however, also has a software area that is ours to update as needed with development software builds and extra stuff as required. Please note that the word 'volume' is used in two different contexts: your "user volume" that you create and share yourself and the system's volumes such as the HEASARC's data volume, which you can also choose to mount. One may prefer to name the "user volumes" with the word "folder" or something to avoid confusion, because they are mounted in different places within the container. III. Files, groups and sharingIn the Home dashboard is Under Files, you can create your own "User volumes" and see what "Data volumes" are accessible (you cannot create your own). You can also browse files through this interface. Under Groups, you can create groups. This is an example, showing the HEASARC software user group as well as others. For each group, it shows you the "Shared Files" (e.g., "user volumes"), the "Shared Data Volumes" (data volumes), and "Shared Compute Images". As you can see, the HEASARC software user group has access to several different things. The Image itself is the instance of Linux plus all the required libraries and software builds already ready to go. The headata volume is where the HEASARCs data holdings will be. You can create your own groups, add your own shared files/volumes, and invite your collaborators to join your group. This way, you and your group can share data that are not available to anybody else. IV. Moving files in and outIn the Jupyter Lab interface, the file navigator on the left side lets you download (right click) and upload (button on top) files. You can also from within your Jupyter Lab use the shell to 'scp' or 'git pull' from external sites that are publicly visible. Thirdly, outside of JupyterLab, the SciServer web site has a Files tab where you can manage the contents of your user volumes. (Note that on occasion, within the container's JupyterLab session, I've found that I got a permission denied trying to remove a file that is listed as owned and writable by the "idies" user I'm running under in the container. In this case, I found I could delete it outside the container in this SciServer->Files tab. This may be because the user volume is mounted with a different set of permissions not visible within the container.) V. SoftwareHEASoftThe image currently has HEASoft 6.30.1:
This is the HEASoft installation you get by default (see your .bashrc). This image requires SciServer admins to update. But you'll also see that you have access to
which we can write to ourselves. If you need it, we can install a development version of HEASoft. (But it takes some time.) Then you'd change your HEADAS to point to that version of HEASoft. Or we can add other software you might need if you cannot install it yourself. CALDB is currently set to the archive's calibration area, which will be kept up to date:
Other mission software is installed in individual conda environments. You can then use conda to activate and de-activate different environments. When you first log in, you will be in the default environment, currently called "(heasoft)". Note that the conda environments are all writeable by the idies user and located in your home directory. This allows you to add software to your Ciao or Fermitools environments as needed. But note that it also allows you to mess up your builds of those packages by mistake, so if this happens, you will need to create a fresh new image. CiaoTo use the Ciao package, you activate the environment and
then run the Ciao
initialization script FermitoolsAs for Ciao, to initialize the Fermitools environment: XMM SAS
XMM SAS also now has its own conda environment, which can be activated with:
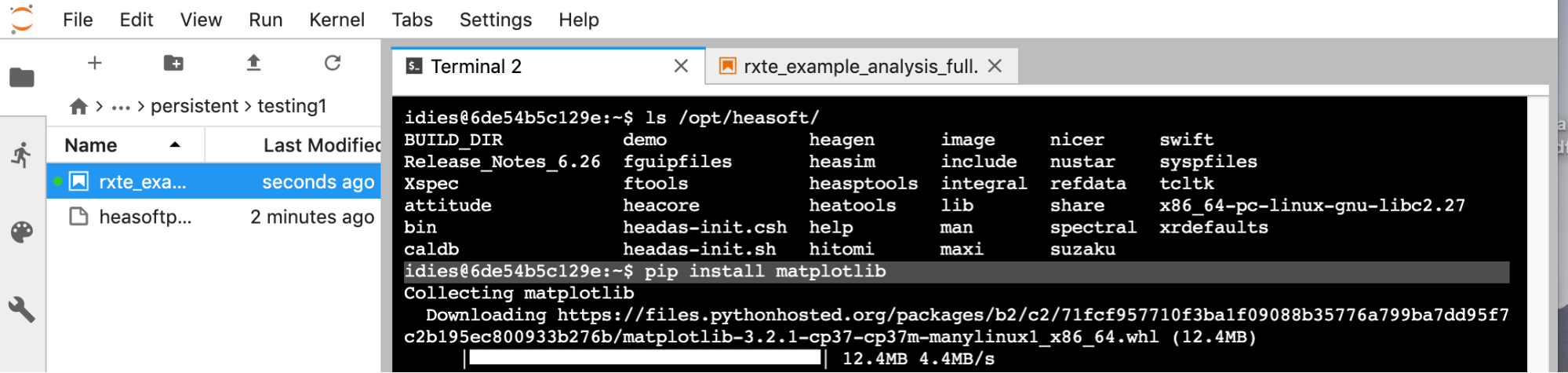
Additional softwareThe software environment is a work in progress. You may need additional libraries. E.g., for additional machine-learning packages, you can:
Alternatively, you can upload your own software and install it. If it uses the distutils, you can run:
If the code doesn't have an install script, then you have to do a bit more. On your desktop, you could add the code location to your PYTHONPATH, but on SciServer, it's difficult to change the environment that the JupyterLab session uses. More straightforwardly, you can to add your code directory to the path inside each notebook using, e.g.,
VI. Test an example notebookWe have several example notebooks in the HEASARC data volume, so if you mount this to your container, then you will find it at:
Make yourself a workspace in your persistent area:
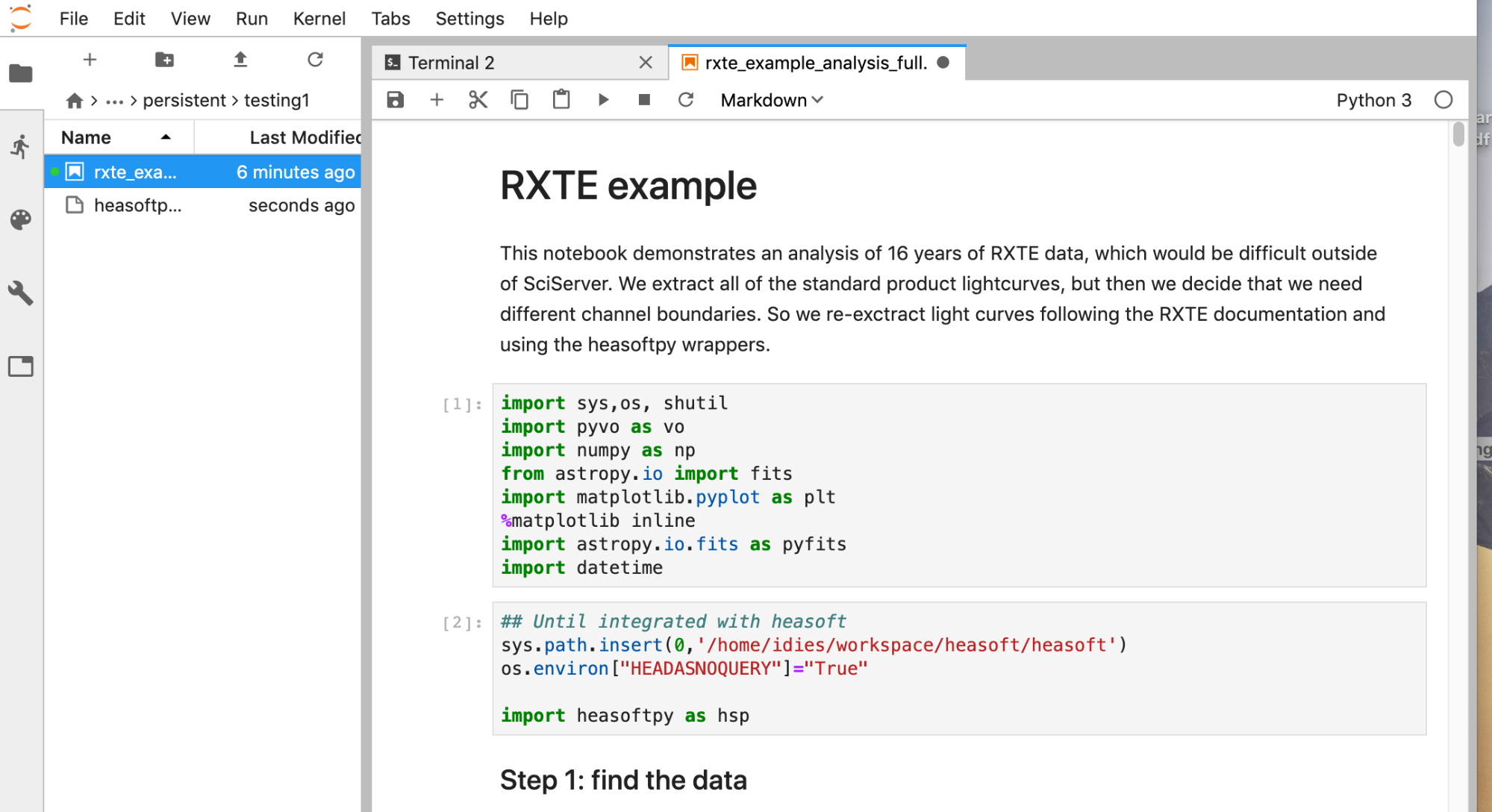
Get a copy of one of our notebooks to try out:
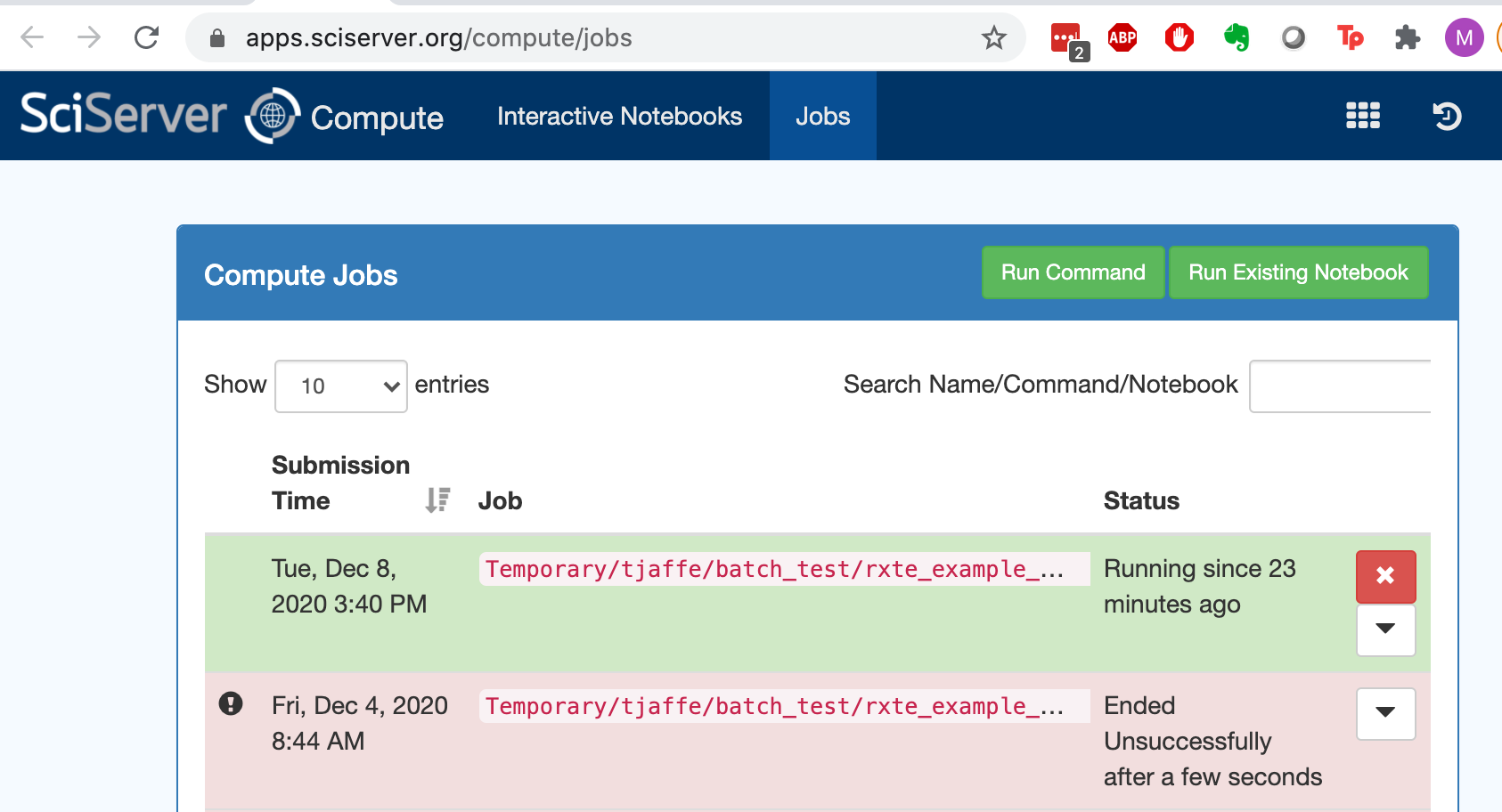
(You can also do this with the navigation sidebar, where you can right click and copy and paste files.) Then in the sidebar on the left, navigate to that directory and double-click on the name of the notebook to open it. Our cookbooks directory also contains other useful notebooks such as the NAVO workshop notebooks describing how to browse and fetch data with VO interfaces, and the HEASARC PyXspec notebooks. Note: for notebooks that are not developed explicitly for this platform, the default Python kernel will not be set correctly. To change it, click on the current kernel, e.g., "Python 3", at the upper right and switch it to "(heasoft)". Otherwise, the necessary libraries may not be available. Starting with version 6.30.1, the image uses python version 3.8. We hope to expand this into a large set of executable analysis threads written by us and our instrument teams. We also encourage our users who have useful notebooks to share them with the community by sending them to us to place in the contrib area. VII. Data discovery with HEASARC toolsThe usual ways of discovering HEASARC data with our Browse and Xamin tools have not yet been integrated seamlessly into SciServer. For Xamin, when you have a list of files or observations you are interested in, there is now an option to generate a simple list starting with the path /FTP that you can then copy-paste into SciServer as needed. Those paths will work on SciServer within your container when you use the HEASARC image and mount the HEASARC data volume. For W3Browse, you can get a download script and then edit that script into a simple file list starting /FTP. (Option pending.) Alternatively, we invite you to explore the Python possibilities. There's a data access notebook in the cookbooks directory with a few examples, one of which is using PyVO. This is a powerful new way to explore not just HEASARC data but data from any VO-compliant archive. See NAVO's collection of notebook tutorials for generic use cases. In the RXTE notebook mentioned above is an example of how to get a list of observations and construct a file list with knowledge of the RXTE archive structure. We hope to provide some Python wrappers for making this more straightforward. VIII. BatchYou can submit a notebook that you have tested interactively to the batch for processing. The batch service is called Compute Jobs (while Compute is interactive) under the main menus. There are a couple of things to be aware of, though. Firstly, we are using the non-standard Python kernel. Set up your Jupyter notebook to use the correct "(Heasarc)" kernel before submitting it to the batch. Submitting to the batch starts with a process a bit like setting up a new container. You have to select the compute image and the volumes that you want to mount. If it starts successfully, it will look something like this: IX. NotesHelpIf you encounter any issues running code or accessing the data, please contact the HEASARC help desk. If your issue is not related to the HEASARC setup specifically, then you can email the SciServer helpdesk. Click on your username at the top right of your dashboard and select Help from the dropdown menu. If you could use some additional software that you cannot install yourself, you can also ask at the HEASARC help desk if we can install it for you. LimitsA summary of users' limitations on SciServer:
See also https://www.sciserver.org/support/policies/. User ContributionsOne of the benefits of SciServer is how easy it makes to share data, code, results, etc. among collaborators. But you can also contribute them to the community of HEASARC@SciServer users. If you have things that you think would be generally useful, place them in a user volume that you can then share with us (e.g., share with user tjaffe), and we'll take a look at whether it would be appropriate to include it on the HEASARC volume for all to use. We also have a notebooks repository on GitHub to which you can contribute through submitting issues to request changes, pull requests to contribute, etc. Likewise for the PyXspec Jupyter notebooks repo. Chandra analysisNote that one cannot use some Ciao tools with the archive itself as the input path, since it will expect to be able to write to the data directories. I.e.,
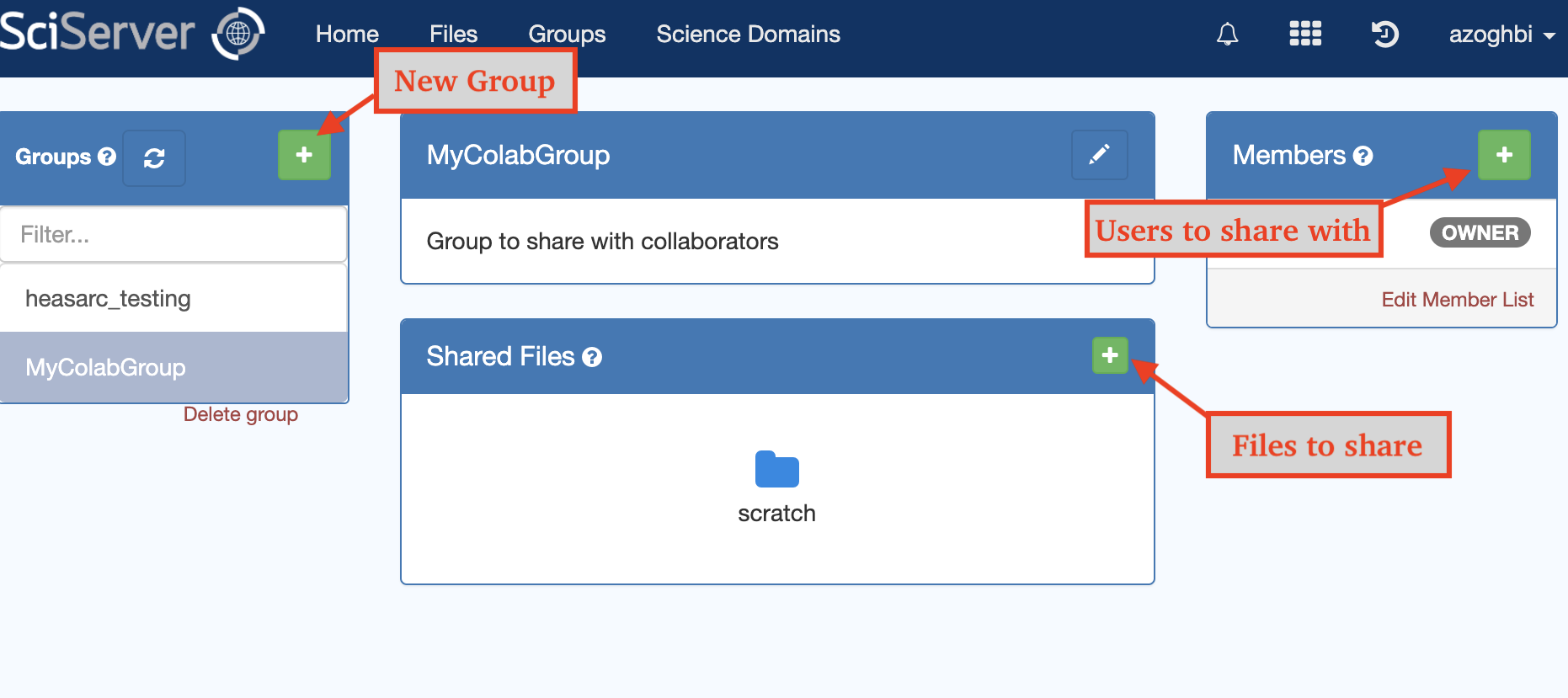
will result in an error about the read-only file system. You will have to copy the input data directory to your own workspace (temporary or persistent as appropriate; see above). XMM analysisLike Chandra analysis, you must copy the input data from the main archive into your own workspace, as it will expect to write to those directories. This is under development, so please send us any issues. Mission-Specific GuidesMiscellaneous
Known Issues
HEASARC Home | Observatories | Archive | Calibration | Software | Tools | Students/Teachers/Public |